Gene
KWMTBOMO08729 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007912
Annotation
PREDICTED:_retinol_dehydrogenase_12-like_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 1.977
Sequence
CDS
ATGGTGTCGGCTCCGGTGATTTATGCAGCAGTGCCCATCTCAGTAGTAGTGGCGATTGGATTAATACGTAAACTGAGATCCAGAAATTGGGGCAAATGTACTAGTAATACATGTCTGAGAGGCAAAACCTTTCTGATAACCGGTGCAAACAGTGGTATCGGACTTGAAACAGCTAAAGCCTTGGTTAAAAGAAAAGCCCGCGTTATCTTTGCATGCAGAGACATTGCAAAGGCGAAAGAAGTTATTGCCGCAATACGAGAGGAACAGCCAAATGGTGGTGAAATGATTCCGATGCAGCTTGACCTTTCATCATTTGAATCTATAGAAAATTTTGTGGATATTATAAAGGCTGGTTTCCATAAAATAGATGTCCTTATAAACAATGCTGGTGTTGTTGTTCCTTTGAATCAAGATGAGAAGACAAAAGAAGGATTTGAAATACATTTTGGTGTGAACCACCTAGGCCACTTTTATCTTACTAATCTTTTGTTAGACCATTTGAAGAGGGCTACTCCAAGTAGAATTGTAATCGTGTCTTCAACGTTACACGAAAAAGGAAAAATCGATTTTGACGACCTAAACTTAAGGTCGGAAATAGAATTAGCGAAAAAGGGCAAAAGCGGTCGTCACAACCCCGGTTATTGCAACTCCAAACTAATGAATGCATACTTTATGAGATCATTAGCTGCAAAATTGAGAAATTCTGGGGTCGATGTCAACGCATGCTGCCCAGGATTCTGTTACACGAACTTATTCAGATATTCGGTAAAATGGTACCACTACATTATGATGGCGCCAATCTTTCTTCTTTTCATGAGATCGGCTAAACAGGGTTCCGAAACAGTATTGTTCTGTGCCACAGATCACAGTCTTGAAGGGAAATCGGGTCGGTTTTACCGCGACTGTGTCGAATACGACACCAAGTATCCATTTGACAGGGCCGTCGAGAACAAACTGTGGGAGGTCAGCGAGGACCTCATCAAAGCCAGGAAGCCGCTGTGA
Protein
MVSAPVIYAAVPISVVVAIGLIRKLRSRNWGKCTSNTCLRGKTFLITGANSGIGLETAKALVKRKARVIFACRDIAKAKEVIAAIREEQPNGGEMIPMQLDLSSFESIENFVDIIKAGFHKIDVLINNAGVVVPLNQDEKTKEGFEIHFGVNHLGHFYLTNLLLDHLKRATPSRIVIVSSTLHEKGKIDFDDLNLRSEIELAKKGKSGRHNPGYCNSKLMNAYFMRSLAAKLRNSGVDVNACCPGFCYTNLFRYSVKWYHYIMMAPIFLLFMRSAKQGSETVLFCATDHSLEGKSGRFYRDCVEYDTKYPFDRAVENKLWEVSEDLIKARKPL
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Belongs to the peptidase C2 family.
Belongs to the peptidase C2 family.
Uniprot
H9JEG5
A0A2A4J2B2
A0A194QNT9
A0A2W1BJ13
A0A3S2NPV7
A0A1Q3G5I0
+ More
A0A212F629 A0A194PWE0 A0A212ERM8 A0A0L7LFN1 S4P0J8 E0VDD8 A0A1B6BX84 A0A0C9QV02 A0A0P4VR99 R4FKX0 A0A2J7Q0L8 A0A067RU66 A0A1B6L1W5 A0A023F7T6 A0A0V0GBR5 A0A224XFS1 A0A0A9YDT0 A0A2R7WJW3 A0A0K8SB02 E2ADL5 A0A026WRM2 A0A158NZW1 A0A151J2K3 A0A195F371 A0A195CNR6 A0A195BLF2 A0A1B6K4T9 E9IT46 A0A151WZX3 F4WIE2 E2C9X5 D6WFR6 A0A087ZX60 A0A232F566 A0A2A3EBL7 A0A0M8ZRS6 A0A3L8DYV4 A0A1W4XJ42 A0A0L7R926 A0A154NYW2 A0A182ML63 A0A2P8YTG4 A0A1B6GF00 A0A1Y1MH66 A0A182Y7X1 A0A1B6FJF3 A0A182SFS5 A0A1B6GC91 A0A084WML5 N6U4K1 Q16QL8 A0NEU8 A0A182UYQ2 A0A182NIW1 W5J866 A0A0T6B2S4 A0A182UJP6 A0A2J7Q0K2 A0A1S4EIL9 A0A3Q0J5E5 A0A1S4EIP8 A0A182PRZ3 A0A1L8E4V6 A0A1S3DB53 A0A182RU56 A0A182I4B7 A0A182X4C2 A0A0P5FHU3 A0A0P5UAJ0 A0A0P6A8F6 A0A1B6GPR4 A0A0P4X3Q9 A0A0P5INM4 A0A0P4WM74 A0A165AAF7 A0A0P5PAF5 A0A0P5CRM4 A0A336KK53 A0A336M7R4 A0A3R7QPT9 A0A0P5E503 A0A1J1IXS7 A0A0P6DX19 A0A1L8E560 A0A1B0D040 A0A0P5K7Q1 A0A0P5T1P7 A0A182FFE2 A0A1S4J8U1 A0A1Q3FLN9 B0W853
A0A212F629 A0A194PWE0 A0A212ERM8 A0A0L7LFN1 S4P0J8 E0VDD8 A0A1B6BX84 A0A0C9QV02 A0A0P4VR99 R4FKX0 A0A2J7Q0L8 A0A067RU66 A0A1B6L1W5 A0A023F7T6 A0A0V0GBR5 A0A224XFS1 A0A0A9YDT0 A0A2R7WJW3 A0A0K8SB02 E2ADL5 A0A026WRM2 A0A158NZW1 A0A151J2K3 A0A195F371 A0A195CNR6 A0A195BLF2 A0A1B6K4T9 E9IT46 A0A151WZX3 F4WIE2 E2C9X5 D6WFR6 A0A087ZX60 A0A232F566 A0A2A3EBL7 A0A0M8ZRS6 A0A3L8DYV4 A0A1W4XJ42 A0A0L7R926 A0A154NYW2 A0A182ML63 A0A2P8YTG4 A0A1B6GF00 A0A1Y1MH66 A0A182Y7X1 A0A1B6FJF3 A0A182SFS5 A0A1B6GC91 A0A084WML5 N6U4K1 Q16QL8 A0NEU8 A0A182UYQ2 A0A182NIW1 W5J866 A0A0T6B2S4 A0A182UJP6 A0A2J7Q0K2 A0A1S4EIL9 A0A3Q0J5E5 A0A1S4EIP8 A0A182PRZ3 A0A1L8E4V6 A0A1S3DB53 A0A182RU56 A0A182I4B7 A0A182X4C2 A0A0P5FHU3 A0A0P5UAJ0 A0A0P6A8F6 A0A1B6GPR4 A0A0P4X3Q9 A0A0P5INM4 A0A0P4WM74 A0A165AAF7 A0A0P5PAF5 A0A0P5CRM4 A0A336KK53 A0A336M7R4 A0A3R7QPT9 A0A0P5E503 A0A1J1IXS7 A0A0P6DX19 A0A1L8E560 A0A1B0D040 A0A0P5K7Q1 A0A0P5T1P7 A0A182FFE2 A0A1S4J8U1 A0A1Q3FLN9 B0W853
Pubmed
EMBL
BABH01003057
NWSH01003932
PCG65664.1
KQ461195
KPJ06640.1
KZ150012
+ More
PZC75062.1 RSAL01000014 RVE53240.1 GEYN01000191 JAV44798.1 AGBW02010081 OWR49192.1 KQ459596 KPI95450.1 AGBW02013012 OWR44101.1 JTDY01001300 KOB74267.1 GAIX01007669 JAA84891.1 DS235073 EEB11394.1 GEDC01031733 JAS05565.1 GBYB01007584 JAG77351.1 GDKW01001665 JAI54930.1 ACPB03001482 GAHY01001663 JAA75847.1 NEVH01019965 PNF22121.1 KK852415 KDR24355.1 GEBQ01022264 JAT17713.1 GBBI01001410 JAC17302.1 GECL01001330 JAP04794.1 GFTR01005100 JAW11326.1 GBHO01013808 JAG29796.1 KK854957 PTY19937.1 GBRD01015359 JAG50467.1 GL438820 EFN68377.1 KK107139 EZA57749.1 ADTU01005049 KQ980404 KYN16218.1 KQ981856 KYN34627.1 KQ977580 KYN01744.1 KQ976440 KYM86515.1 GECU01001251 JAT06456.1 GL765434 EFZ16373.1 KQ982629 KYQ53426.1 GL888175 EGI66073.1 GL453904 EFN75273.1 KQ971319 EEZ99585.1 NNAY01000983 OXU25620.1 KZ288292 PBC29080.1 KQ435883 KOX69795.1 QOIP01000003 RLU25129.1 KQ414628 KOC67370.1 KQ434775 KZC04060.1 AXCM01000273 PYGN01000369 PSN47539.1 GECZ01008760 JAS61009.1 GEZM01031355 JAV84931.1 GECZ01028757 GECZ01019464 GECZ01003948 JAS41012.1 JAS50305.1 JAS65821.1 GECZ01016086 GECZ01009711 JAS53683.1 JAS60058.1 ATLV01024482 KE525352 KFB51459.1 APGK01049519 KB741156 KB632169 ENN73512.1 ERL89431.1 CH477744 EAT36698.1 AAAB01008960 EAU76619.2 ADMH02001882 ETN60662.1 LJIG01016080 KRT81676.1 PNF22118.1 GFDF01000420 JAV13664.1 APCN01002300 GDIQ01255060 JAJ96664.1 GDIP01153150 GDIQ01153386 GDIP01115754 GDIP01115753 GDIP01041163 GDIQ01036817 JAK98339.1 JAL87961.1 GDIP01214393 GDIP01214392 GDIP01140051 GDIP01045869 GDIQ01075665 JAM57846.1 JAN19072.1 GECZ01005345 JAS64424.1 GDIP01254926 JAI68475.1 GDIQ01211265 JAK40460.1 GDIP01254927 JAI68474.1 LRGB01000642 KZS17371.1 GDIQ01131356 JAL20370.1 GDIP01167631 JAJ55771.1 UFQS01000608 UFQT01000608 SSX05314.1 SSX25675.1 UFQS01000513 UFQT01000513 SSX04509.1 SSX24873.1 QCYY01000029 ROT86048.1 GDIP01164997 JAJ58405.1 CVRI01000064 CRL05075.1 GDIQ01071018 JAN23719.1 GFDF01000389 JAV13695.1 AJVK01009809 GDIQ01188220 JAK63505.1 GDIP01132300 JAL71414.1 GFDL01006632 JAV28413.1 DS231857 EDS38678.1
PZC75062.1 RSAL01000014 RVE53240.1 GEYN01000191 JAV44798.1 AGBW02010081 OWR49192.1 KQ459596 KPI95450.1 AGBW02013012 OWR44101.1 JTDY01001300 KOB74267.1 GAIX01007669 JAA84891.1 DS235073 EEB11394.1 GEDC01031733 JAS05565.1 GBYB01007584 JAG77351.1 GDKW01001665 JAI54930.1 ACPB03001482 GAHY01001663 JAA75847.1 NEVH01019965 PNF22121.1 KK852415 KDR24355.1 GEBQ01022264 JAT17713.1 GBBI01001410 JAC17302.1 GECL01001330 JAP04794.1 GFTR01005100 JAW11326.1 GBHO01013808 JAG29796.1 KK854957 PTY19937.1 GBRD01015359 JAG50467.1 GL438820 EFN68377.1 KK107139 EZA57749.1 ADTU01005049 KQ980404 KYN16218.1 KQ981856 KYN34627.1 KQ977580 KYN01744.1 KQ976440 KYM86515.1 GECU01001251 JAT06456.1 GL765434 EFZ16373.1 KQ982629 KYQ53426.1 GL888175 EGI66073.1 GL453904 EFN75273.1 KQ971319 EEZ99585.1 NNAY01000983 OXU25620.1 KZ288292 PBC29080.1 KQ435883 KOX69795.1 QOIP01000003 RLU25129.1 KQ414628 KOC67370.1 KQ434775 KZC04060.1 AXCM01000273 PYGN01000369 PSN47539.1 GECZ01008760 JAS61009.1 GEZM01031355 JAV84931.1 GECZ01028757 GECZ01019464 GECZ01003948 JAS41012.1 JAS50305.1 JAS65821.1 GECZ01016086 GECZ01009711 JAS53683.1 JAS60058.1 ATLV01024482 KE525352 KFB51459.1 APGK01049519 KB741156 KB632169 ENN73512.1 ERL89431.1 CH477744 EAT36698.1 AAAB01008960 EAU76619.2 ADMH02001882 ETN60662.1 LJIG01016080 KRT81676.1 PNF22118.1 GFDF01000420 JAV13664.1 APCN01002300 GDIQ01255060 JAJ96664.1 GDIP01153150 GDIQ01153386 GDIP01115754 GDIP01115753 GDIP01041163 GDIQ01036817 JAK98339.1 JAL87961.1 GDIP01214393 GDIP01214392 GDIP01140051 GDIP01045869 GDIQ01075665 JAM57846.1 JAN19072.1 GECZ01005345 JAS64424.1 GDIP01254926 JAI68475.1 GDIQ01211265 JAK40460.1 GDIP01254927 JAI68474.1 LRGB01000642 KZS17371.1 GDIQ01131356 JAL20370.1 GDIP01167631 JAJ55771.1 UFQS01000608 UFQT01000608 SSX05314.1 SSX25675.1 UFQS01000513 UFQT01000513 SSX04509.1 SSX24873.1 QCYY01000029 ROT86048.1 GDIP01164997 JAJ58405.1 CVRI01000064 CRL05075.1 GDIQ01071018 JAN23719.1 GFDF01000389 JAV13695.1 AJVK01009809 GDIQ01188220 JAK63505.1 GDIP01132300 JAL71414.1 GFDL01006632 JAV28413.1 DS231857 EDS38678.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000283053
UP000007151
UP000053268
+ More
UP000037510 UP000009046 UP000015103 UP000235965 UP000027135 UP000000311 UP000053097 UP000005205 UP000078492 UP000078541 UP000078542 UP000078540 UP000075809 UP000007755 UP000008237 UP000007266 UP000005203 UP000215335 UP000242457 UP000053105 UP000279307 UP000192223 UP000053825 UP000076502 UP000075883 UP000245037 UP000076408 UP000075901 UP000030765 UP000019118 UP000030742 UP000008820 UP000007062 UP000075903 UP000075884 UP000000673 UP000075902 UP000079169 UP000075885 UP000075900 UP000075840 UP000076407 UP000076858 UP000283509 UP000183832 UP000092462 UP000069272 UP000002320
UP000037510 UP000009046 UP000015103 UP000235965 UP000027135 UP000000311 UP000053097 UP000005205 UP000078492 UP000078541 UP000078542 UP000078540 UP000075809 UP000007755 UP000008237 UP000007266 UP000005203 UP000215335 UP000242457 UP000053105 UP000279307 UP000192223 UP000053825 UP000076502 UP000075883 UP000245037 UP000076408 UP000075901 UP000030765 UP000019118 UP000030742 UP000008820 UP000007062 UP000075903 UP000075884 UP000000673 UP000075902 UP000079169 UP000075885 UP000075900 UP000075840 UP000076407 UP000076858 UP000283509 UP000183832 UP000092462 UP000069272 UP000002320
PRIDE
Pfam
Interpro
IPR002347
SDR_fam
+ More
IPR036291 NAD(P)-bd_dom_sf
IPR022682 Calpain_domain_III
IPR000169 Pept_cys_AS
IPR018247 EF_Hand_1_Ca_BS
IPR036213 Calpain_III_sf
IPR033883 C2_III
IPR038765 Papain-like_cys_pep_sf
IPR022684 Calpain_cysteine_protease
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR022683 Calpain_III
IPR001300 Peptidase_C2_calpain_cat
IPR019013 Vma21
IPR036982 Deoxyhypusine_synthase_sf
IPR002773 Deoxyhypusine_synthase
IPR029035 DHS-like_NAD/FAD-binding_dom
IPR006722 Sedlin
IPR011012 Longin-like_dom_sf
IPR036291 NAD(P)-bd_dom_sf
IPR022682 Calpain_domain_III
IPR000169 Pept_cys_AS
IPR018247 EF_Hand_1_Ca_BS
IPR036213 Calpain_III_sf
IPR033883 C2_III
IPR038765 Papain-like_cys_pep_sf
IPR022684 Calpain_cysteine_protease
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR022683 Calpain_III
IPR001300 Peptidase_C2_calpain_cat
IPR019013 Vma21
IPR036982 Deoxyhypusine_synthase_sf
IPR002773 Deoxyhypusine_synthase
IPR029035 DHS-like_NAD/FAD-binding_dom
IPR006722 Sedlin
IPR011012 Longin-like_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JEG5
A0A2A4J2B2
A0A194QNT9
A0A2W1BJ13
A0A3S2NPV7
A0A1Q3G5I0
+ More
A0A212F629 A0A194PWE0 A0A212ERM8 A0A0L7LFN1 S4P0J8 E0VDD8 A0A1B6BX84 A0A0C9QV02 A0A0P4VR99 R4FKX0 A0A2J7Q0L8 A0A067RU66 A0A1B6L1W5 A0A023F7T6 A0A0V0GBR5 A0A224XFS1 A0A0A9YDT0 A0A2R7WJW3 A0A0K8SB02 E2ADL5 A0A026WRM2 A0A158NZW1 A0A151J2K3 A0A195F371 A0A195CNR6 A0A195BLF2 A0A1B6K4T9 E9IT46 A0A151WZX3 F4WIE2 E2C9X5 D6WFR6 A0A087ZX60 A0A232F566 A0A2A3EBL7 A0A0M8ZRS6 A0A3L8DYV4 A0A1W4XJ42 A0A0L7R926 A0A154NYW2 A0A182ML63 A0A2P8YTG4 A0A1B6GF00 A0A1Y1MH66 A0A182Y7X1 A0A1B6FJF3 A0A182SFS5 A0A1B6GC91 A0A084WML5 N6U4K1 Q16QL8 A0NEU8 A0A182UYQ2 A0A182NIW1 W5J866 A0A0T6B2S4 A0A182UJP6 A0A2J7Q0K2 A0A1S4EIL9 A0A3Q0J5E5 A0A1S4EIP8 A0A182PRZ3 A0A1L8E4V6 A0A1S3DB53 A0A182RU56 A0A182I4B7 A0A182X4C2 A0A0P5FHU3 A0A0P5UAJ0 A0A0P6A8F6 A0A1B6GPR4 A0A0P4X3Q9 A0A0P5INM4 A0A0P4WM74 A0A165AAF7 A0A0P5PAF5 A0A0P5CRM4 A0A336KK53 A0A336M7R4 A0A3R7QPT9 A0A0P5E503 A0A1J1IXS7 A0A0P6DX19 A0A1L8E560 A0A1B0D040 A0A0P5K7Q1 A0A0P5T1P7 A0A182FFE2 A0A1S4J8U1 A0A1Q3FLN9 B0W853
A0A212F629 A0A194PWE0 A0A212ERM8 A0A0L7LFN1 S4P0J8 E0VDD8 A0A1B6BX84 A0A0C9QV02 A0A0P4VR99 R4FKX0 A0A2J7Q0L8 A0A067RU66 A0A1B6L1W5 A0A023F7T6 A0A0V0GBR5 A0A224XFS1 A0A0A9YDT0 A0A2R7WJW3 A0A0K8SB02 E2ADL5 A0A026WRM2 A0A158NZW1 A0A151J2K3 A0A195F371 A0A195CNR6 A0A195BLF2 A0A1B6K4T9 E9IT46 A0A151WZX3 F4WIE2 E2C9X5 D6WFR6 A0A087ZX60 A0A232F566 A0A2A3EBL7 A0A0M8ZRS6 A0A3L8DYV4 A0A1W4XJ42 A0A0L7R926 A0A154NYW2 A0A182ML63 A0A2P8YTG4 A0A1B6GF00 A0A1Y1MH66 A0A182Y7X1 A0A1B6FJF3 A0A182SFS5 A0A1B6GC91 A0A084WML5 N6U4K1 Q16QL8 A0NEU8 A0A182UYQ2 A0A182NIW1 W5J866 A0A0T6B2S4 A0A182UJP6 A0A2J7Q0K2 A0A1S4EIL9 A0A3Q0J5E5 A0A1S4EIP8 A0A182PRZ3 A0A1L8E4V6 A0A1S3DB53 A0A182RU56 A0A182I4B7 A0A182X4C2 A0A0P5FHU3 A0A0P5UAJ0 A0A0P6A8F6 A0A1B6GPR4 A0A0P4X3Q9 A0A0P5INM4 A0A0P4WM74 A0A165AAF7 A0A0P5PAF5 A0A0P5CRM4 A0A336KK53 A0A336M7R4 A0A3R7QPT9 A0A0P5E503 A0A1J1IXS7 A0A0P6DX19 A0A1L8E560 A0A1B0D040 A0A0P5K7Q1 A0A0P5T1P7 A0A182FFE2 A0A1S4J8U1 A0A1Q3FLN9 B0W853
PDB
3RD5
E-value=1.54474e-31,
Score=339
Ontologies
PANTHER
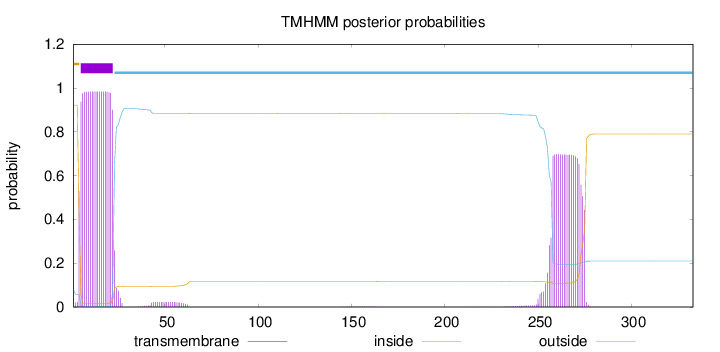
Topology
Length:
333
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
32.17362
Exp number, first 60 AAs:
19.02017
Total prob of N-in:
0.07936
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 22
inside
23 - 333
Population Genetic Test Statistics
Pi
40.821656
Theta
35.833527
Tajima's D
-1.702733
CLR
2.39602
CSRT
0.0362981850907455
Interpretation
Uncertain
