Gene
KWMTBOMO08723
Pre Gene Modal
BGIBMGA007914
Annotation
PREDICTED:_nuclear_hormone_receptor_FTZ-F1_beta_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 2.437
Sequence
CDS
ATGTTTGGGGGAATACGACACAGGGAAATCGACGACCAAATCTGCCTGCTGATCAACAGTTGGTGCGAGCTGCTTGTGCTTTCTTGCTGCTACCGCGGTGTCAGCACCCCGGGTGAAGTGAGAGTCGGAGGCGGTAGAGGCATCACTCTACATCAAAGTGCCAAACTCGGTTTGACGCCGTGTATAGAACGGATGCTGAGCTTTACTGATCACTTGAGGAGGCTACGCGTCGACAGATACGAATACGTTGCTCTTAAAGTCATCGTTCTATTGACTTCAGACGCGCCTGAGCTACGCGAGTCCGAAAAAGTTCGAGCCTCGCAAGAGAAAGCTTTGGCAGCCCTGCAAGCTTACATAGCGACTCATTCGCCCGGAACACCCGCTAAGTTTGGTGAACTCTTGCTCCGGATTCCGGAACTGCAGAGAACATGTCAGTTCTTCATGCAAGTCGGTAAAGAAATGTTAAATCCCGCCAACAAGAATAAAGATGGCGACGGACAGAGCTTCAACCTCCTTATGGAATTACTGAGAGGTGACCATTAA
Protein
MFGGIRHREIDDQICLLINSWCELLVLSCCYRGVSTPGEVRVGGGRGITLHQSAKLGLTPCIERMLSFTDHLRRLRVDRYEYVALKVIVLLTSDAPELRESEKVRASQEKALAALQAYIATHSPGTPAKFGELLLRIPELQRTCQFFMQVGKEMLNPANKNKDGDGQSFNLLMELLRGDH
Summary
Similarity
Belongs to the nuclear hormone receptor family.
Uniprot
O96041
H9JEG7
A0A2H1WBZ8
A0A2W1BQP5
A0A3S2LGD2
A0A2A4K6H4
+ More
A0A212FLD1 A0A194QM90 A0A194PQ55 A0A182UIK0 A0A182GP53 A0A1B6HNN8 A0A1B6MIJ8 A0A1B6F5Y0 A0A1S4H106 A0A1W4WWF8 A0A1Q3F3U4 A0A1Q3F3U7 A0A1S4H1R5 G8Z9Y8 A0A2H8TI65 D2A6H1 A0A1Y1L3R0 J9K0T6 A0A2S2PA12 A0A2M4BL49 A0A2S2Q9G4 A0A182QPF0 A0A182MLA4 A0A182L6Q8 A0A2M4BK44 A0A2M4ADE5 A0A182UXK3 A0A182GLJ0 A0A182GKK1 A0A182YFW1 A0A182VUQ4 A0A2M3Z672 W5J5Q5 A0A182HG72 A0A1S4EYC9 Q7QGB9 Q17LN1 A0A182P2S0 A0A1Q3F3N6 A0A1Q3F3X0 A0A182XDK5 A0A182RG74 A0A0C9R5H7 B0WAZ1 A0A182NEN2 A0A2M4BFC9 A0A2M4CVP5 A0A0T6B6T1 A0A2M4A7C2 A0A182F1V5 A0A2J7R1A4 A0A084VLW8 A0A2J7R1B8 A0A1B0DH33 A0A067R992 A0A2M4A873 A0A0V0G7N3 A0A224XL16 A0A182IXG7 A0A023F481 A0A069DWE2 A0A1B6DMQ3 T1HFZ6 A0A1J1I9C9 E0VMS4 A0A2Z2XT94 A0A2R7W7P1 E9J400 A0A0P4W677 A0A0L7R3C0 A0A195D3S7 E2BMH6 A0A2A3E9R0 A0A195EQJ4 A0A310SB35 A0A151XB37 A0A336K8C1 K7IWC6 F4WKA7 A0A195BS21 A0A026WUY5 A0A195DHV7 A0A158NQJ9 A0A232FK25 A0A0J7L4X9 A0A3L8E4L0 T1PB10 E9H1N6 E1ZXF8 A0A0P4WEM0 A0A0K8T693 A0A0K8T695 A0A2P8YJG0 A0A1A9X064
A0A212FLD1 A0A194QM90 A0A194PQ55 A0A182UIK0 A0A182GP53 A0A1B6HNN8 A0A1B6MIJ8 A0A1B6F5Y0 A0A1S4H106 A0A1W4WWF8 A0A1Q3F3U4 A0A1Q3F3U7 A0A1S4H1R5 G8Z9Y8 A0A2H8TI65 D2A6H1 A0A1Y1L3R0 J9K0T6 A0A2S2PA12 A0A2M4BL49 A0A2S2Q9G4 A0A182QPF0 A0A182MLA4 A0A182L6Q8 A0A2M4BK44 A0A2M4ADE5 A0A182UXK3 A0A182GLJ0 A0A182GKK1 A0A182YFW1 A0A182VUQ4 A0A2M3Z672 W5J5Q5 A0A182HG72 A0A1S4EYC9 Q7QGB9 Q17LN1 A0A182P2S0 A0A1Q3F3N6 A0A1Q3F3X0 A0A182XDK5 A0A182RG74 A0A0C9R5H7 B0WAZ1 A0A182NEN2 A0A2M4BFC9 A0A2M4CVP5 A0A0T6B6T1 A0A2M4A7C2 A0A182F1V5 A0A2J7R1A4 A0A084VLW8 A0A2J7R1B8 A0A1B0DH33 A0A067R992 A0A2M4A873 A0A0V0G7N3 A0A224XL16 A0A182IXG7 A0A023F481 A0A069DWE2 A0A1B6DMQ3 T1HFZ6 A0A1J1I9C9 E0VMS4 A0A2Z2XT94 A0A2R7W7P1 E9J400 A0A0P4W677 A0A0L7R3C0 A0A195D3S7 E2BMH6 A0A2A3E9R0 A0A195EQJ4 A0A310SB35 A0A151XB37 A0A336K8C1 K7IWC6 F4WKA7 A0A195BS21 A0A026WUY5 A0A195DHV7 A0A158NQJ9 A0A232FK25 A0A0J7L4X9 A0A3L8E4L0 T1PB10 E9H1N6 E1ZXF8 A0A0P4WEM0 A0A0K8T693 A0A0K8T695 A0A2P8YJG0 A0A1A9X064
Pubmed
EMBL
AB005660
BAA34123.1
BABH01003065
BABH01003066
ODYU01007583
SOQ50476.1
+ More
KZ150012 PZC75060.1 RSAL01000014 RVE53238.1 NWSH01000128 PCG79272.1 AGBW02007796 OWR54555.1 KQ461195 KPJ06637.1 KQ459596 KPI95447.1 JXUM01003132 KQ560153 KXJ84149.1 GECU01031409 JAS76297.1 GEBQ01004187 JAT35790.1 GECZ01024149 JAS45620.1 AAAB01008835 GFDL01012809 JAV22236.1 GFDL01012815 JAV22230.1 GU479987 ADK46871.1 GFXV01001153 MBW12958.1 KQ971348 EFA05671.1 GEZM01071539 GEZM01071538 JAV65717.1 ABLF02024872 ABLF02024873 GGMR01013631 MBY26250.1 GGFJ01004540 MBW53681.1 GGMS01005161 MBY74364.1 AXCN02000847 AXCM01000060 GGFJ01004284 MBW53425.1 GGFK01005493 MBW38814.1 JXUM01072215 KQ562708 KXJ75294.1 JXUM01001078 JXUM01001079 GGFM01003268 MBW24019.1 ADMH02002158 ETN58200.1 APCN01004189 EAA05800.4 CH477214 EAT47581.1 GFDL01012898 JAV22147.1 GFDL01012791 JAV22254.1 GBYB01008032 JAG77799.1 DS231876 EDS41949.1 GGFJ01002586 MBW51727.1 GGFL01005234 MBW69412.1 LJIG01009579 KRT82797.1 GGFK01003358 MBW36679.1 NEVH01008208 PNF34620.1 ATLV01014570 KE524975 KFB38962.1 PNF34622.1 AJVK01015319 AJVK01015320 KK852804 KDR16159.1 GGFK01003507 MBW36828.1 GECL01002110 JAP04014.1 GFTR01007705 JAW08721.1 GBBI01002888 JAC15824.1 GBGD01000723 JAC88166.1 GEDC01017966 GEDC01010345 JAS19332.1 JAS26953.1 ACPB03006526 CVRI01000044 CRK96819.1 DS235324 EEB14680.1 KY817190 ATX63078.1 KK854423 PTY15744.1 GL768069 EFZ12441.1 GDRN01068968 GDRN01068966 JAI64117.1 KQ414663 KOC65370.1 KQ976885 KYN07547.1 GL449233 EFN83085.1 KZ288311 PBC28513.1 KQ982021 KYN30550.1 KQ761976 OAD56427.1 KQ982335 KYQ57508.1 UFQS01000121 UFQT01000121 SSX00062.1 SSX20442.1 GL888199 EGI65331.1 KQ976419 KYM89309.1 KK107087 EZA59860.1 KQ980824 KYN12483.1 ADTU01023357 NNAY01000110 OXU30890.1 LBMM01000709 KMQ97676.1 QOIP01000001 RLU26918.1 KA645335 AFP59964.1 GL732583 EFX74471.1 GL435038 EFN74135.1 GDRN01068967 JAI64118.1 GBRD01004779 GBRD01004778 GBRD01004776 GBRD01004774 GDHC01017357 JAG61042.1 JAQ01272.1 GBRD01004780 GBRD01004777 GBRD01004775 GBRD01004773 GDHC01017583 JAG61044.1 JAQ01046.1 PYGN01000551 PSN44390.1
KZ150012 PZC75060.1 RSAL01000014 RVE53238.1 NWSH01000128 PCG79272.1 AGBW02007796 OWR54555.1 KQ461195 KPJ06637.1 KQ459596 KPI95447.1 JXUM01003132 KQ560153 KXJ84149.1 GECU01031409 JAS76297.1 GEBQ01004187 JAT35790.1 GECZ01024149 JAS45620.1 AAAB01008835 GFDL01012809 JAV22236.1 GFDL01012815 JAV22230.1 GU479987 ADK46871.1 GFXV01001153 MBW12958.1 KQ971348 EFA05671.1 GEZM01071539 GEZM01071538 JAV65717.1 ABLF02024872 ABLF02024873 GGMR01013631 MBY26250.1 GGFJ01004540 MBW53681.1 GGMS01005161 MBY74364.1 AXCN02000847 AXCM01000060 GGFJ01004284 MBW53425.1 GGFK01005493 MBW38814.1 JXUM01072215 KQ562708 KXJ75294.1 JXUM01001078 JXUM01001079 GGFM01003268 MBW24019.1 ADMH02002158 ETN58200.1 APCN01004189 EAA05800.4 CH477214 EAT47581.1 GFDL01012898 JAV22147.1 GFDL01012791 JAV22254.1 GBYB01008032 JAG77799.1 DS231876 EDS41949.1 GGFJ01002586 MBW51727.1 GGFL01005234 MBW69412.1 LJIG01009579 KRT82797.1 GGFK01003358 MBW36679.1 NEVH01008208 PNF34620.1 ATLV01014570 KE524975 KFB38962.1 PNF34622.1 AJVK01015319 AJVK01015320 KK852804 KDR16159.1 GGFK01003507 MBW36828.1 GECL01002110 JAP04014.1 GFTR01007705 JAW08721.1 GBBI01002888 JAC15824.1 GBGD01000723 JAC88166.1 GEDC01017966 GEDC01010345 JAS19332.1 JAS26953.1 ACPB03006526 CVRI01000044 CRK96819.1 DS235324 EEB14680.1 KY817190 ATX63078.1 KK854423 PTY15744.1 GL768069 EFZ12441.1 GDRN01068968 GDRN01068966 JAI64117.1 KQ414663 KOC65370.1 KQ976885 KYN07547.1 GL449233 EFN83085.1 KZ288311 PBC28513.1 KQ982021 KYN30550.1 KQ761976 OAD56427.1 KQ982335 KYQ57508.1 UFQS01000121 UFQT01000121 SSX00062.1 SSX20442.1 GL888199 EGI65331.1 KQ976419 KYM89309.1 KK107087 EZA59860.1 KQ980824 KYN12483.1 ADTU01023357 NNAY01000110 OXU30890.1 LBMM01000709 KMQ97676.1 QOIP01000001 RLU26918.1 KA645335 AFP59964.1 GL732583 EFX74471.1 GL435038 EFN74135.1 GDRN01068967 JAI64118.1 GBRD01004779 GBRD01004778 GBRD01004776 GBRD01004774 GDHC01017357 JAG61042.1 JAQ01272.1 GBRD01004780 GBRD01004777 GBRD01004775 GBRD01004773 GDHC01017583 JAG61044.1 JAQ01046.1 PYGN01000551 PSN44390.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000075902 UP000069940 UP000249989 UP000192223 UP000007266 UP000007819 UP000075886 UP000075883 UP000075882 UP000075903 UP000076408 UP000075920 UP000000673 UP000075840 UP000007062 UP000008820 UP000075885 UP000076407 UP000075900 UP000002320 UP000075884 UP000069272 UP000235965 UP000030765 UP000092462 UP000027135 UP000075880 UP000015103 UP000183832 UP000009046 UP000053825 UP000078542 UP000008237 UP000242457 UP000078541 UP000075809 UP000002358 UP000007755 UP000078540 UP000053097 UP000078492 UP000005205 UP000215335 UP000036403 UP000279307 UP000000305 UP000000311 UP000245037 UP000091820
UP000075902 UP000069940 UP000249989 UP000192223 UP000007266 UP000007819 UP000075886 UP000075883 UP000075882 UP000075903 UP000076408 UP000075920 UP000000673 UP000075840 UP000007062 UP000008820 UP000075885 UP000076407 UP000075900 UP000002320 UP000075884 UP000069272 UP000235965 UP000030765 UP000092462 UP000027135 UP000075880 UP000015103 UP000183832 UP000009046 UP000053825 UP000078542 UP000008237 UP000242457 UP000078541 UP000075809 UP000002358 UP000007755 UP000078540 UP000053097 UP000078492 UP000005205 UP000215335 UP000036403 UP000279307 UP000000305 UP000000311 UP000245037 UP000091820
Interpro
SUPFAM
SSF48508
SSF48508
Gene 3D
ProteinModelPortal
O96041
H9JEG7
A0A2H1WBZ8
A0A2W1BQP5
A0A3S2LGD2
A0A2A4K6H4
+ More
A0A212FLD1 A0A194QM90 A0A194PQ55 A0A182UIK0 A0A182GP53 A0A1B6HNN8 A0A1B6MIJ8 A0A1B6F5Y0 A0A1S4H106 A0A1W4WWF8 A0A1Q3F3U4 A0A1Q3F3U7 A0A1S4H1R5 G8Z9Y8 A0A2H8TI65 D2A6H1 A0A1Y1L3R0 J9K0T6 A0A2S2PA12 A0A2M4BL49 A0A2S2Q9G4 A0A182QPF0 A0A182MLA4 A0A182L6Q8 A0A2M4BK44 A0A2M4ADE5 A0A182UXK3 A0A182GLJ0 A0A182GKK1 A0A182YFW1 A0A182VUQ4 A0A2M3Z672 W5J5Q5 A0A182HG72 A0A1S4EYC9 Q7QGB9 Q17LN1 A0A182P2S0 A0A1Q3F3N6 A0A1Q3F3X0 A0A182XDK5 A0A182RG74 A0A0C9R5H7 B0WAZ1 A0A182NEN2 A0A2M4BFC9 A0A2M4CVP5 A0A0T6B6T1 A0A2M4A7C2 A0A182F1V5 A0A2J7R1A4 A0A084VLW8 A0A2J7R1B8 A0A1B0DH33 A0A067R992 A0A2M4A873 A0A0V0G7N3 A0A224XL16 A0A182IXG7 A0A023F481 A0A069DWE2 A0A1B6DMQ3 T1HFZ6 A0A1J1I9C9 E0VMS4 A0A2Z2XT94 A0A2R7W7P1 E9J400 A0A0P4W677 A0A0L7R3C0 A0A195D3S7 E2BMH6 A0A2A3E9R0 A0A195EQJ4 A0A310SB35 A0A151XB37 A0A336K8C1 K7IWC6 F4WKA7 A0A195BS21 A0A026WUY5 A0A195DHV7 A0A158NQJ9 A0A232FK25 A0A0J7L4X9 A0A3L8E4L0 T1PB10 E9H1N6 E1ZXF8 A0A0P4WEM0 A0A0K8T693 A0A0K8T695 A0A2P8YJG0 A0A1A9X064
A0A212FLD1 A0A194QM90 A0A194PQ55 A0A182UIK0 A0A182GP53 A0A1B6HNN8 A0A1B6MIJ8 A0A1B6F5Y0 A0A1S4H106 A0A1W4WWF8 A0A1Q3F3U4 A0A1Q3F3U7 A0A1S4H1R5 G8Z9Y8 A0A2H8TI65 D2A6H1 A0A1Y1L3R0 J9K0T6 A0A2S2PA12 A0A2M4BL49 A0A2S2Q9G4 A0A182QPF0 A0A182MLA4 A0A182L6Q8 A0A2M4BK44 A0A2M4ADE5 A0A182UXK3 A0A182GLJ0 A0A182GKK1 A0A182YFW1 A0A182VUQ4 A0A2M3Z672 W5J5Q5 A0A182HG72 A0A1S4EYC9 Q7QGB9 Q17LN1 A0A182P2S0 A0A1Q3F3N6 A0A1Q3F3X0 A0A182XDK5 A0A182RG74 A0A0C9R5H7 B0WAZ1 A0A182NEN2 A0A2M4BFC9 A0A2M4CVP5 A0A0T6B6T1 A0A2M4A7C2 A0A182F1V5 A0A2J7R1A4 A0A084VLW8 A0A2J7R1B8 A0A1B0DH33 A0A067R992 A0A2M4A873 A0A0V0G7N3 A0A224XL16 A0A182IXG7 A0A023F481 A0A069DWE2 A0A1B6DMQ3 T1HFZ6 A0A1J1I9C9 E0VMS4 A0A2Z2XT94 A0A2R7W7P1 E9J400 A0A0P4W677 A0A0L7R3C0 A0A195D3S7 E2BMH6 A0A2A3E9R0 A0A195EQJ4 A0A310SB35 A0A151XB37 A0A336K8C1 K7IWC6 F4WKA7 A0A195BS21 A0A026WUY5 A0A195DHV7 A0A158NQJ9 A0A232FK25 A0A0J7L4X9 A0A3L8E4L0 T1PB10 E9H1N6 E1ZXF8 A0A0P4WEM0 A0A0K8T693 A0A0K8T695 A0A2P8YJG0 A0A1A9X064
PDB
3CJW
E-value=3.26906e-16,
Score=203
Ontologies
GO
PANTHER
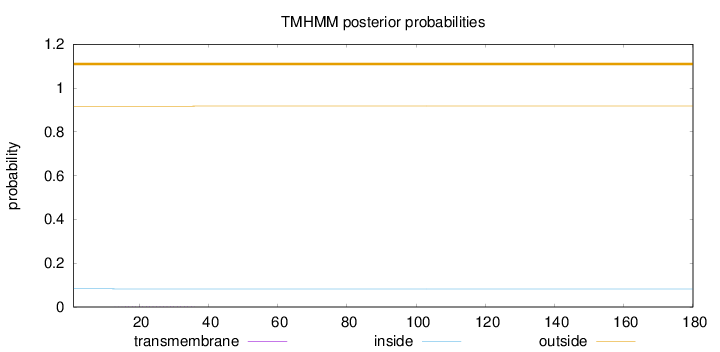
Topology
Subcellular location
Nucleus
Length:
180
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0348
Exp number, first 60 AAs:
0.03284
Total prob of N-in:
0.08330
outside
1 - 180
Population Genetic Test Statistics
Pi
129.468194
Theta
143.369186
Tajima's D
-0.339075
CLR
5.956896
CSRT
0.275386230688466
Interpretation
Uncertain
