Gene
KWMTBOMO08722
Pre Gene Modal
BGIBMGA007915
Annotation
hatching_enzyme-like_protein_precursor_[Bombyx_mori]
Full name
Metalloendopeptidase
Location in the cell
Cytoplasmic Reliability : 1.522
Sequence
CDS
ATGTTGCGTATTGCAGTATTAGTTACTATCGTCTGCTCCGCGTTGGCCACGCCCGCTGTCGTGAGGACTAGAGAGGAAATACAGGCTTTCAGAAGCTTCCTGGAAAGCATCAAATCCGGCGATGGGAGTCAGTTCGCCGCGAGAACCATGACCAATCCTCTGGCCAACCCTGAGGAGAACAGCGGCAAATTCCAAGGAGATATTGTCCTAGACGACTTCATGATTGAAATCCTACTCAGGCAGTACGCAATGGGGCGTAACGCGTACACGTGGCCCAATACTAAGTGGCCGAACAACACCGTTGTTTGGGAATTTGGAGAGGGGGAATTTGGTCCTCGTCAGCAAGCCGCCATCGAGGAGGGGATCAGAGACATCGAAAAACACACATGCCTCAAGTTCCGTTACCGCGAGGCTGGAGACACAGCCTTTGTCAGACTCACCGGCGGAGCTGGTGGATGTTATGCCCACGTCGGTTACTGGGAATCACGAGGCGTGCACGTGTTGAACCTTGCCCGCAACGAGCCAGGAGTGGGCTGTTTCCGCCACGCCACCATCGTGCACGAGTGGATGCACATCCTCGGCTTCCTCCACATGCAGTCCACCCACAACAGAGACGACTATGTTAAGATCGTCAAAGAAAACATTACGCCTGGTCTTCAGCACAACTTCGACATCTACACACAAGATCTCGTTGGTAACTTAGGCGTTGAATACGACTATGTCAGTTGTCTGCATTACGGACCTTACGCATTCTCACAAAACGGGGAGAAAACTATCGTTGCTCTGCAGGAGCACGAGGGTGTGATGGGTCAGCGTTTGTATGTAACCGACGATGACTGGCTTCGCATCAACAGACATTACAACTGCGAAGGAGCTTGGAACTAA
Protein
MLRIAVLVTIVCSALATPAVVRTREEIQAFRSFLESIKSGDGSQFAARTMTNPLANPEENSGKFQGDIVLDDFMIEILLRQYAMGRNAYTWPNTKWPNNTVVWEFGEGEFGPRQQAAIEEGIRDIEKHTCLKFRYREAGDTAFVRLTGGAGGCYAHVGYWESRGVHVLNLARNEPGVGCFRHATIVHEWMHILGFLHMQSTHNRDDYVKIVKENITPGLQHNFDIYTQDLVGNLGVEYDYVSCLHYGPYAFSQNGEKTIVALQEHEGVMGQRLYVTDDDWLRINRHYNCEGAWN
Summary
Cofactor
Zn(2+)
Feature
chain Metalloendopeptidase
Uniprot
H9JEG8
B5TZ28
G3M4F7
H9JEG9
G3M5H2
X5D7Z9
+ More
G1FJ09 A0A2Z4KH61 A0A2Z4KH36 A0A0K0XQP5 A0A1E1W6X6 A0A3S2NZN5 F1C4B7 A0A212F628 A0A194PRR7 A0A194QNT4 A0A437BS11 A0A0L7L3V4 A0A2W1BN49 A0A2H1WGP1 A0A2A4K5E0 A0A2W1BN55 B5B473 F6K710 A0A2H1WBN7 A0A2A4K5D1 A0A437ASQ8 A0A2A4K6I0 A0A2A4K4U7 A0A2W1BJ38 A0A2W1BP44 A0A2W1BJL4 A0A0K8T4D0 A0A2W1BQ83 A0A3S2NKG8 A0A3S2M7H4 A0A437BSB0 A0A1E1WM51 A0A182GAC7 Q16PS3 A0A1S4FU18 Q16PS1 Q16G58 A0A0A1XQW1 A0A0M4ES32 B0W777 B0W775 B0W776 A0A1Q3FQ05 A0A034WRL2 A0A1I8QC97 A0A0K8UJS9 A0A0A1XJ49 A0A0K8V774 A0A182Q180 A0A182WXS3 A0A034WJI4 B4NY97 B4Q6G4 Q9GTN4 A0A182FVY3 Q9GTN3 A0A0J9TNU9 A0A2C9GRW7 A0A182V0D5 A0A182MQ71 A0A1I8N2C9 A0A182YIV7 I1W6K7 B4JQV2 B3N5Z9 Q9V421 A0A182U9M7 A0A1I8MEV6 A0A1W4VFH3 A0A182SLS2 Q29KV4 B4LV66 A0A182GAC8 B4KLI4 B4MYZ9 A0A2H1WVD0 A0A182KV80 A0A182I5U4 A0A2A4K808 Q7Q209 A0A182X0K4 A0A182UM81 A0A182UCN0 A0A3B0JI07 B3N5Z6 Q16PS6 A0A084WUG9 A0A0K8URH9 W8CA95 A0A0K8UEU3 T1DK26 A0A194PQ49 Q4V4J5 A0A0L0BS59
G1FJ09 A0A2Z4KH61 A0A2Z4KH36 A0A0K0XQP5 A0A1E1W6X6 A0A3S2NZN5 F1C4B7 A0A212F628 A0A194PRR7 A0A194QNT4 A0A437BS11 A0A0L7L3V4 A0A2W1BN49 A0A2H1WGP1 A0A2A4K5E0 A0A2W1BN55 B5B473 F6K710 A0A2H1WBN7 A0A2A4K5D1 A0A437ASQ8 A0A2A4K6I0 A0A2A4K4U7 A0A2W1BJ38 A0A2W1BP44 A0A2W1BJL4 A0A0K8T4D0 A0A2W1BQ83 A0A3S2NKG8 A0A3S2M7H4 A0A437BSB0 A0A1E1WM51 A0A182GAC7 Q16PS3 A0A1S4FU18 Q16PS1 Q16G58 A0A0A1XQW1 A0A0M4ES32 B0W777 B0W775 B0W776 A0A1Q3FQ05 A0A034WRL2 A0A1I8QC97 A0A0K8UJS9 A0A0A1XJ49 A0A0K8V774 A0A182Q180 A0A182WXS3 A0A034WJI4 B4NY97 B4Q6G4 Q9GTN4 A0A182FVY3 Q9GTN3 A0A0J9TNU9 A0A2C9GRW7 A0A182V0D5 A0A182MQ71 A0A1I8N2C9 A0A182YIV7 I1W6K7 B4JQV2 B3N5Z9 Q9V421 A0A182U9M7 A0A1I8MEV6 A0A1W4VFH3 A0A182SLS2 Q29KV4 B4LV66 A0A182GAC8 B4KLI4 B4MYZ9 A0A2H1WVD0 A0A182KV80 A0A182I5U4 A0A2A4K808 Q7Q209 A0A182X0K4 A0A182UM81 A0A182UCN0 A0A3B0JI07 B3N5Z6 Q16PS6 A0A084WUG9 A0A0K8URH9 W8CA95 A0A0K8UEU3 T1DK26 A0A194PQ49 Q4V4J5 A0A0L0BS59
EC Number
3.4.24.-
Pubmed
EMBL
BABH01003066
BABH01003067
BABH01003068
FJ147197
ACI02327.1
JN620366
+ More
AEO80031.1 BABH01003074 BABH01003075 BABH01003076 JN627443 AEO53067.1 KJ184542 KJ642218 AHW50928.1 AID50186.1 JN205047 AEL12457.1 MH119084 AWW91899.1 MH129044 AWW91898.1 KT336227 AKS39779.1 GDQN01008349 JAT82705.1 RSAL01000014 RVE53237.1 HQ199327 ADX30518.1 AGBW02010081 OWR49195.1 KQ459596 KPI95444.1 KQ461195 KPJ06635.1 RVE53234.1 JTDY01003106 KOB70143.1 KZ150012 PZC75054.1 ODYU01008518 SOQ52187.1 NWSH01000128 PCG79279.1 PZC75055.1 EU915282 ACG69475.1 HM357847 AEA76313.1 ODYU01007583 SOQ50478.1 PCG79269.1 RSAL01000960 RVE41020.1 PCG79270.1 PCG79271.1 PZC75059.1 PZC75057.1 PZC75058.1 GBRD01005429 JAG60392.1 PZC75056.1 RVE53233.1 RVE53236.1 RVE53235.1 GDQN01002974 GDQN01002826 JAT88080.1 JAT88228.1 JXUM01008840 KQ560270 KXJ83413.1 CH477774 EAT36349.1 EAT36351.1 CH478328 EAT33220.1 EAT36348.1 GBXI01000975 JAD13317.1 CP012523 ALC39807.1 DS231852 EDS37682.1 EDS37680.1 EDS37681.1 GFDL01005431 JAV29614.1 GAKP01002544 JAC56408.1 GDHF01025365 JAI26949.1 GBXI01003377 JAD10915.1 GDHF01017706 JAI34608.1 AXCN02001809 GAKP01004672 GAKP01004671 JAC54280.1 CM000157 EDW89733.1 CM000361 CM002910 EDX05146.1 KMY90389.1 AF264913 AAG10255.1 AF264914 AAG10256.1 KMY90390.1 APCN01003509 AXCM01005146 BT133482 AFI61834.1 CH916372 EDV99282.1 CH954177 EDV58037.1 AE014134 AAF53471.1 CH379061 EAL33070.1 CH940649 EDW64326.1 JXUM01008841 JXUM01008842 KXJ83414.1 CH933807 EDW12865.1 CH963913 EDW77338.1 ODYU01011338 SOQ57025.1 APCN01003503 NWSH01000058 PCG80078.1 AAAB01008979 EAA13734.4 OUUW01000006 SPP81835.1 EDV58034.1 EAT36346.1 ATLV01027083 KE525423 KFB53863.1 GDHF01023123 GDHF01010372 JAI29191.1 JAI41942.1 GAMC01005961 JAC00595.1 GDHF01027479 JAI24835.1 GAMD01001161 JAB00430.1 KQ459602 KPI93260.1 BT023011 AAY55427.1 JRES01001454 KNC22838.1
AEO80031.1 BABH01003074 BABH01003075 BABH01003076 JN627443 AEO53067.1 KJ184542 KJ642218 AHW50928.1 AID50186.1 JN205047 AEL12457.1 MH119084 AWW91899.1 MH129044 AWW91898.1 KT336227 AKS39779.1 GDQN01008349 JAT82705.1 RSAL01000014 RVE53237.1 HQ199327 ADX30518.1 AGBW02010081 OWR49195.1 KQ459596 KPI95444.1 KQ461195 KPJ06635.1 RVE53234.1 JTDY01003106 KOB70143.1 KZ150012 PZC75054.1 ODYU01008518 SOQ52187.1 NWSH01000128 PCG79279.1 PZC75055.1 EU915282 ACG69475.1 HM357847 AEA76313.1 ODYU01007583 SOQ50478.1 PCG79269.1 RSAL01000960 RVE41020.1 PCG79270.1 PCG79271.1 PZC75059.1 PZC75057.1 PZC75058.1 GBRD01005429 JAG60392.1 PZC75056.1 RVE53233.1 RVE53236.1 RVE53235.1 GDQN01002974 GDQN01002826 JAT88080.1 JAT88228.1 JXUM01008840 KQ560270 KXJ83413.1 CH477774 EAT36349.1 EAT36351.1 CH478328 EAT33220.1 EAT36348.1 GBXI01000975 JAD13317.1 CP012523 ALC39807.1 DS231852 EDS37682.1 EDS37680.1 EDS37681.1 GFDL01005431 JAV29614.1 GAKP01002544 JAC56408.1 GDHF01025365 JAI26949.1 GBXI01003377 JAD10915.1 GDHF01017706 JAI34608.1 AXCN02001809 GAKP01004672 GAKP01004671 JAC54280.1 CM000157 EDW89733.1 CM000361 CM002910 EDX05146.1 KMY90389.1 AF264913 AAG10255.1 AF264914 AAG10256.1 KMY90390.1 APCN01003509 AXCM01005146 BT133482 AFI61834.1 CH916372 EDV99282.1 CH954177 EDV58037.1 AE014134 AAF53471.1 CH379061 EAL33070.1 CH940649 EDW64326.1 JXUM01008841 JXUM01008842 KXJ83414.1 CH933807 EDW12865.1 CH963913 EDW77338.1 ODYU01011338 SOQ57025.1 APCN01003503 NWSH01000058 PCG80078.1 AAAB01008979 EAA13734.4 OUUW01000006 SPP81835.1 EDV58034.1 EAT36346.1 ATLV01027083 KE525423 KFB53863.1 GDHF01023123 GDHF01010372 JAI29191.1 JAI41942.1 GAMC01005961 JAC00595.1 GDHF01027479 JAI24835.1 GAMD01001161 JAB00430.1 KQ459602 KPI93260.1 BT023011 AAY55427.1 JRES01001454 KNC22838.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000218220 UP000069940 UP000249989 UP000008820 UP000092553 UP000002320 UP000095300 UP000075886 UP000076407 UP000002282 UP000000304 UP000069272 UP000075840 UP000075903 UP000075883 UP000095301 UP000076408 UP000001070 UP000008711 UP000000803 UP000075902 UP000192221 UP000075901 UP000001819 UP000008792 UP000009192 UP000007798 UP000075882 UP000007062 UP000268350 UP000030765 UP000037069
UP000218220 UP000069940 UP000249989 UP000008820 UP000092553 UP000002320 UP000095300 UP000075886 UP000076407 UP000002282 UP000000304 UP000069272 UP000075840 UP000075903 UP000075883 UP000095301 UP000076408 UP000001070 UP000008711 UP000000803 UP000075902 UP000192221 UP000075901 UP000001819 UP000008792 UP000009192 UP000007798 UP000075882 UP000007062 UP000268350 UP000030765 UP000037069
Pfam
PF01400 Astacin
Interpro
Gene 3D
ProteinModelPortal
H9JEG8
B5TZ28
G3M4F7
H9JEG9
G3M5H2
X5D7Z9
+ More
G1FJ09 A0A2Z4KH61 A0A2Z4KH36 A0A0K0XQP5 A0A1E1W6X6 A0A3S2NZN5 F1C4B7 A0A212F628 A0A194PRR7 A0A194QNT4 A0A437BS11 A0A0L7L3V4 A0A2W1BN49 A0A2H1WGP1 A0A2A4K5E0 A0A2W1BN55 B5B473 F6K710 A0A2H1WBN7 A0A2A4K5D1 A0A437ASQ8 A0A2A4K6I0 A0A2A4K4U7 A0A2W1BJ38 A0A2W1BP44 A0A2W1BJL4 A0A0K8T4D0 A0A2W1BQ83 A0A3S2NKG8 A0A3S2M7H4 A0A437BSB0 A0A1E1WM51 A0A182GAC7 Q16PS3 A0A1S4FU18 Q16PS1 Q16G58 A0A0A1XQW1 A0A0M4ES32 B0W777 B0W775 B0W776 A0A1Q3FQ05 A0A034WRL2 A0A1I8QC97 A0A0K8UJS9 A0A0A1XJ49 A0A0K8V774 A0A182Q180 A0A182WXS3 A0A034WJI4 B4NY97 B4Q6G4 Q9GTN4 A0A182FVY3 Q9GTN3 A0A0J9TNU9 A0A2C9GRW7 A0A182V0D5 A0A182MQ71 A0A1I8N2C9 A0A182YIV7 I1W6K7 B4JQV2 B3N5Z9 Q9V421 A0A182U9M7 A0A1I8MEV6 A0A1W4VFH3 A0A182SLS2 Q29KV4 B4LV66 A0A182GAC8 B4KLI4 B4MYZ9 A0A2H1WVD0 A0A182KV80 A0A182I5U4 A0A2A4K808 Q7Q209 A0A182X0K4 A0A182UM81 A0A182UCN0 A0A3B0JI07 B3N5Z6 Q16PS6 A0A084WUG9 A0A0K8URH9 W8CA95 A0A0K8UEU3 T1DK26 A0A194PQ49 Q4V4J5 A0A0L0BS59
G1FJ09 A0A2Z4KH61 A0A2Z4KH36 A0A0K0XQP5 A0A1E1W6X6 A0A3S2NZN5 F1C4B7 A0A212F628 A0A194PRR7 A0A194QNT4 A0A437BS11 A0A0L7L3V4 A0A2W1BN49 A0A2H1WGP1 A0A2A4K5E0 A0A2W1BN55 B5B473 F6K710 A0A2H1WBN7 A0A2A4K5D1 A0A437ASQ8 A0A2A4K6I0 A0A2A4K4U7 A0A2W1BJ38 A0A2W1BP44 A0A2W1BJL4 A0A0K8T4D0 A0A2W1BQ83 A0A3S2NKG8 A0A3S2M7H4 A0A437BSB0 A0A1E1WM51 A0A182GAC7 Q16PS3 A0A1S4FU18 Q16PS1 Q16G58 A0A0A1XQW1 A0A0M4ES32 B0W777 B0W775 B0W776 A0A1Q3FQ05 A0A034WRL2 A0A1I8QC97 A0A0K8UJS9 A0A0A1XJ49 A0A0K8V774 A0A182Q180 A0A182WXS3 A0A034WJI4 B4NY97 B4Q6G4 Q9GTN4 A0A182FVY3 Q9GTN3 A0A0J9TNU9 A0A2C9GRW7 A0A182V0D5 A0A182MQ71 A0A1I8N2C9 A0A182YIV7 I1W6K7 B4JQV2 B3N5Z9 Q9V421 A0A182U9M7 A0A1I8MEV6 A0A1W4VFH3 A0A182SLS2 Q29KV4 B4LV66 A0A182GAC8 B4KLI4 B4MYZ9 A0A2H1WVD0 A0A182KV80 A0A182I5U4 A0A2A4K808 Q7Q209 A0A182X0K4 A0A182UM81 A0A182UCN0 A0A3B0JI07 B3N5Z6 Q16PS6 A0A084WUG9 A0A0K8URH9 W8CA95 A0A0K8UEU3 T1DK26 A0A194PQ49 Q4V4J5 A0A0L0BS59
PDB
4GWM
E-value=4.65113e-29,
Score=317
Ontologies
GO
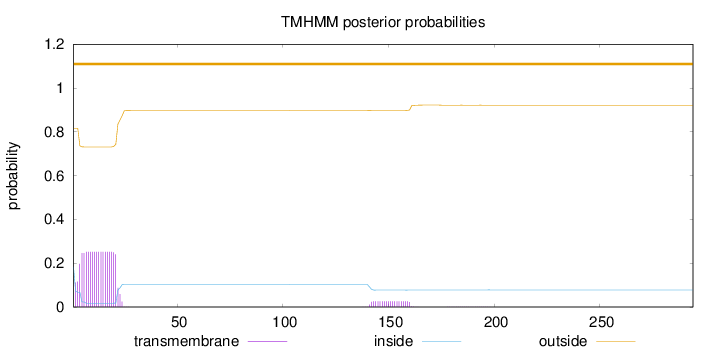
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.953347
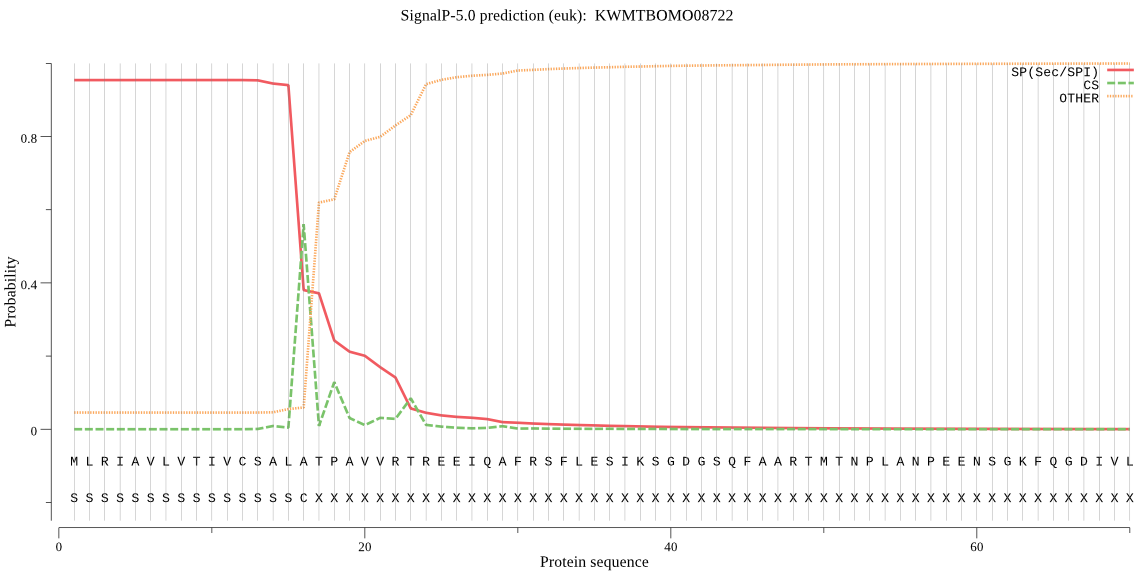
Length:
294
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.36910000000001
Exp number, first 60 AAs:
4.85244
Total prob of N-in:
0.18356
outside
1 - 294
Population Genetic Test Statistics
Pi
186.76176
Theta
159.24408
Tajima's D
-0.739891
CLR
0.926136
CSRT
0.191440427978601
Interpretation
Uncertain
