Gene
KWMTBOMO08721
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.269
Sequence
CDS
ATGGAGACTCGACATAATAAATTACGTATAGGACCGCTGCCCGAGGGCGATCGTCGGGGCACGTCTGTAGCTGGCCACGGGCGTGACAGCATGCTAACCAGTGGCTCCCGGACCACCGGCTGTACGGGAGTCGGGGACGCGGGGGGGAGGGGAAAGTCCTCAAGAGCTACAGCCGAGGCGATAAGTGGGAGTAAATCAGCAACGCCAGCTGAGGATGTTTCGCAGGAGACATACGTCACCCAACCTGCGACCACCAAAGCTGGTAAACCAAGAGTGCGCATGAAATGGAGTGAAGATATTAACCTATTTATCATGCGCACATATTTCTACATTACCAAACTAGAAACGGACAAGACACAACGGAAAAGACTTCATGAACGATTCCGACATCGGTACCCTAGTTTAGACGTATCGTCACAAAGAATAGCAGACCAAAGAAGAGTTATACTAAGAAATAGACTTCTATCAGAACAAACTATTGACAACATTAAAAAATCAGTCGCAGAGCAACTTAAACAAGAAGAACAGGACCATTTCAATACATCTAGACATAATGTATCCACCACACAACAACCAACTATGCCAGTCATCTTTGAAAAAGATCCCGCAAACTCCGACTATTATTCAACATCAATCGATATGGTCACAAATCAACAACTTTCAACTACAATAAGACATCAATCATGTGAAGTTTCGACACAAACAAATACGCTCCCAGAGTGGACTACACTTTTACAATCTGCAGAACAAGATAAATCTCTCGAAACAGCTATCACTAACGAACTAGCACAAAAACTTAACACGCAGCTTGAAACAACACTAATTCAATATTCAGGTACAGACCCTACATCTCGTCCCAAACTACCTCGATTAAAAGAGAATAAAACTTTATACGACTTAGTTGCAGTTTTTAACAAAGAAATTCTCGGTAAACACTTCACGATTGAATCTGACATCTGTGACATACAGTCACTGGTCTACTGCAGTGCTTTGGTTATTTCTGAGCATTTAGGATATAAAATCAGAAATAATCTACTATCACCCGAAAACCGTACACGTAAATCGCGAAAACCCTGTTGGCAATTTAGAATCGAAAATGATATTAAGAGACTGCGAGCTGAAATAGGTAGATTAACACAATACATTAACAACCATAATAACAGGTCTGATCGATTAATTAAAGCAGTTGAACAAATTTTTAGGAACACAAGAATTCATTCATCATATGAGAATAATAATACCAGGCCAGAAGAATTTTTGGACACATTAAAACAGAAATTAGCACTTAAATCACAGAGACTAAGGTAG
Protein
METRHNKLRIGPLPEGDRRGTSVAGHGRDSMLTSGSRTTGCTGVGDAGGRGKSSRATAEAISGSKSATPAEDVSQETYVTQPATTKAGKPRVRMKWSEDINLFIMRTYFYITKLETDKTQRKRLHERFRHRYPSLDVSSQRIADQRRVILRNRLLSEQTIDNIKKSVAEQLKQEEQDHFNTSRHNVSTTQQPTMPVIFEKDPANSDYYSTSIDMVTNQQLSTTIRHQSCEVSTQTNTLPEWTTLLQSAEQDKSLETAITNELAQKLNTQLETTLIQYSGTDPTSRPKLPRLKENKTLYDLVAVFNKEILGKHFTIESDICDIQSLVYCSALVISEHLGYKIRNNLLSPENRTRKSRKPCWQFRIENDIKRLRAEIGRLTQYINNHNNRSDRLIKAVEQIFRNTRIHSSYENNNTRPEEFLDTLKQKLALKSQRLR
Summary
Uniprot
Pubmed
EMBL
L08889
AAA92147.1
GAMC01018219
JAB88336.1
GAMC01018221
JAB88334.1
+ More
KZ150118 PZC73245.1 GEZM01018924 JAV89960.1 ODYU01006945 SOQ49255.1 GAMC01016897 JAB89658.1 GEZM01018922 JAV89961.1 AFYH01106869 AB097127 BAC82595.1 RSAL01001347 RVE40825.1 UFQT01000728 SSX26770.1 KZ150426 PZC70922.1 KQ972532 KXZ75876.1 AAWZ02033877 DS235750 EEB16295.1 DS235366 EEB15300.1 DS235861 EEB19180.1 EEB15312.1 DS235859 EEB19129.1 DS235060 EEB11140.1
KZ150118 PZC73245.1 GEZM01018924 JAV89960.1 ODYU01006945 SOQ49255.1 GAMC01016897 JAB89658.1 GEZM01018922 JAV89961.1 AFYH01106869 AB097127 BAC82595.1 RSAL01001347 RVE40825.1 UFQT01000728 SSX26770.1 KZ150426 PZC70922.1 KQ972532 KXZ75876.1 AAWZ02033877 DS235750 EEB16295.1 DS235366 EEB15300.1 DS235861 EEB19180.1 EEB15312.1 DS235859 EEB19129.1 DS235060 EEB11140.1
Proteomes
PRIDE
Pfam
PF00078 RVT_1
ProteinModelPortal
Ontologies
GO
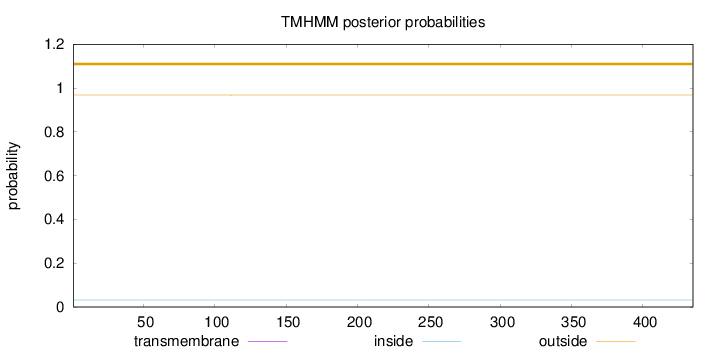
Topology
Length:
435
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00383
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03214
outside
1 - 435
Population Genetic Test Statistics
Pi
29.687565
Theta
14.095705
Tajima's D
3.313944
CLR
0
CSRT
0.990550472476376
Interpretation
Uncertain
