Gene
KWMTBOMO08717
Pre Gene Modal
BGIBMGA007918
Annotation
Mitochondrial_cardiolipin_hydrolase_[Papilio_machaon]
Full name
Mitochondrial cardiolipin hydrolase
Alternative Name
Phospholipase D6 homolog
Location in the cell
PlasmaMembrane Reliability : 1.715
Sequence
CDS
ATGGACAGATTTTTACACTATTTAGCTTCACCCAAGTACAGTCTTGATGTATGTATTTACGTTTTCACCAACTCTGATATTGCGAATGTGTTACTAAAACTACACTACAAAGGTATTAAAATCCGCATTATCATAGATGCCGACATGGCATACTGTACAGGTTCAAATTTGAGAAGAATGGAGCGTCAAGGCATACCTGTACGCTGGATGAAGTCTACAAATCTAATGCACCACAAGTTCTGCATAATAGATGCAGTGAATATAGATGATGTACACCCTTTAGTAATAGCAGGGTCATTGAACTGGACCAATCAGGCACTTTGCGGAAACTGGGAGAATGTTCTGGTGACATCACAAGCTGATCTTGTAAACCAGTTTAAGACTGAATTTGAGAAACTTTGGTTACAGTTTACACCAATAACCAGTTAA
Protein
MDRFLHYLASPKYSLDVCIYVFTNSDIANVLLKLHYKGIKIRIIIDADMAYCTGSNLRRMERQGIPVRWMKSTNLMHHKFCIIDAVNIDDVHPLVIAGSLNWTNQALCGNWENVLVTSQADLVNQFKTEFEKLWLQFTPITS
Summary
Description
Plays a critical role in PIWI-interacting RNA (piRNA) biogenesis (By similarity). piRNAs provide essential protection against the activity of mobile genetic elements. piRNA-mediated transposon silencing is thus critical for maintaining genome stability. Backbone-non-specific, single strand-specific nuclease, cleaving either RNA or DNA substrates with similar affinity (By similarity). Produces 5' phosphate and 3' hydroxyl termini, suggesting it could directly participate in the processing of primary piRNA transcripts (By similarity). Has been proposed to act as a cardiolipin hydrolase to generate phosphatidic acid at mitochondrial surface. Although it cannot be excluded that it can act as a phospholipase in some circumstances, this activity could not be confirmed (By similarity).
Subunit
Homodimer.
Similarity
Belongs to the phospholipase D family. MitoPLD/Zucchini subfamily.
Keywords
Endonuclease
Hydrolase
Lipid degradation
Lipid metabolism
Meiosis
Membrane
Metal-binding
Mitochondrion
Mitochondrion outer membrane
Nuclease
Transmembrane
Transmembrane helix
Feature
chain Mitochondrial cardiolipin hydrolase
Uniprot
H9JEH1
A0A194QM63
A0A2A4K4V7
A0A194PQ50
A0A2W1BJ03
A0A0L7LJI0
+ More
A0A2H1WGJ8 A0A2J8AF98 A0A1B6GEV7 A0A067RHN9 D8UHV0 A0A1I8MWS3 A0A1B6KCR5 A0A1B6H873 A0A2K3D149 A8IW99 A0A1I8PG51 W8AYK0 K7WRZ7 B4IE38 K7WRZ0 K7WPV2 A0A1W1ELN7 A0AP53 K7WPV0 K7WKG9 A0A0A1WEX7 K7WRY1 K7XHQ2 K7WRZ4 H2ZTC7 K7XHQ0 K7X4V0 K7WPU5 K7WPV8 K7WKI0 R7UPE4 K7XHP5 K7WKH3 K7WKG6 K7WPV6 K7WRY6 A0A023GD06 A0A2J7R8C9 A0A1E1XDP6 K7XHP1 K7WKH6 A0A034VDP1 A0A1A8APG1 C1EET0 A0A2S1PS15 K7X4U0 A0A3S1B8F8 T1JLA2 A0A2S1PRW4 A0A2E5HM73 A0A2T7Q0Z7 T2MA50 A0A1W1ELM3 A0A3Q1H821 A0A1S3K043 F6TQM8 A0A3B4VN71 A0A3B4YFF5 A0A3Q3KC05 A0A1B6DNY1 A0A232FI96 A7SU88 A0A1S3Q697 A0A3B5ARB5 A0A1S3Q608 A0A1S3Q907 A0A131YZG8 A0A3M6UA87 A0A1E1XQA7 U3KH83 A0A3Q3RKB0 A0A2C9K659 A0A3P8Z8K9 T1EF80 A0A3P9ANA1 A0A1A9X7N3 K7X570 A0A2U1JD42 A0A2T9Z0I0 A0A2S1PRV2 A0A3P9HJQ8 H2LEV7 A0A3P9M3E3 A0A2S1PRU9 A0A210QQ12 A0A3Q1CKK2 A0A1W4VAK5 A0A401PGT5 A0A224YMV2 A0A3L8Q451 G3Q753 A0A3P8Z8M3 K7WKW4
A0A2H1WGJ8 A0A2J8AF98 A0A1B6GEV7 A0A067RHN9 D8UHV0 A0A1I8MWS3 A0A1B6KCR5 A0A1B6H873 A0A2K3D149 A8IW99 A0A1I8PG51 W8AYK0 K7WRZ7 B4IE38 K7WRZ0 K7WPV2 A0A1W1ELN7 A0AP53 K7WPV0 K7WKG9 A0A0A1WEX7 K7WRY1 K7XHQ2 K7WRZ4 H2ZTC7 K7XHQ0 K7X4V0 K7WPU5 K7WPV8 K7WKI0 R7UPE4 K7XHP5 K7WKH3 K7WKG6 K7WPV6 K7WRY6 A0A023GD06 A0A2J7R8C9 A0A1E1XDP6 K7XHP1 K7WKH6 A0A034VDP1 A0A1A8APG1 C1EET0 A0A2S1PS15 K7X4U0 A0A3S1B8F8 T1JLA2 A0A2S1PRW4 A0A2E5HM73 A0A2T7Q0Z7 T2MA50 A0A1W1ELM3 A0A3Q1H821 A0A1S3K043 F6TQM8 A0A3B4VN71 A0A3B4YFF5 A0A3Q3KC05 A0A1B6DNY1 A0A232FI96 A7SU88 A0A1S3Q697 A0A3B5ARB5 A0A1S3Q608 A0A1S3Q907 A0A131YZG8 A0A3M6UA87 A0A1E1XQA7 U3KH83 A0A3Q3RKB0 A0A2C9K659 A0A3P8Z8K9 T1EF80 A0A3P9ANA1 A0A1A9X7N3 K7X570 A0A2U1JD42 A0A2T9Z0I0 A0A2S1PRV2 A0A3P9HJQ8 H2LEV7 A0A3P9M3E3 A0A2S1PRU9 A0A210QQ12 A0A3Q1CKK2 A0A1W4VAK5 A0A401PGT5 A0A224YMV2 A0A3L8Q451 G3Q753 A0A3P8Z8M3 K7WKW4
EC Number
3.1.-.-
Pubmed
19121390
26354079
28756777
26227816
29294063
24845553
+ More
20616280 25315136 17932292 24495485 24361206 17994087 16951084 22936249 25830018 9215903 23254933 28503490 25348373 19359590 29337314 24065732 17495919 28648823 17615350 26830274 30382153 29209593 15562597 25069045 29764946 17554307 28812685 30297745 28797301 30282656
20616280 25315136 17932292 24495485 24361206 17994087 16951084 22936249 25830018 9215903 23254933 28503490 25348373 19359590 29337314 24065732 17495919 28648823 17615350 26830274 30382153 29209593 15562597 25069045 29764946 17554307 28812685 30297745 28797301 30282656
EMBL
BABH01003077
KQ461195
KPJ06633.1
NWSH01000128
PCG79277.1
KQ459596
+ More
KPI95442.1 KZ150012 PZC75052.1 JTDY01000843 KOB75693.1 ODYU01008518 SOQ52189.1 PGGS01000036 PNH11184.1 GECZ01008801 JAS60968.1 KK852464 KDR23312.1 GL378409 EFJ40702.1 GEBQ01030751 JAT09226.1 GECU01036892 JAS70814.1 CM008974 PNW74265.1 DS496125 GAMC01015305 GAMC01015303 JAB91252.1 JX656776 AFX62716.1 CH480831 EDW45865.1 JX656766 AFX62706.1 JX656764 JX656765 AFX62704.1 LT674447 SIP63003.1 AM294441 CM000361 CM002910 CAL26384.1 EDX04750.1 KMY89848.1 JX656759 AFX62699.1 JX656760 JX656762 AFX62702.1 GBXI01017267 JAC97024.1 JX656756 AFX62696.1 JX656763 JX656773 JX656775 AFX62713.1 JX656771 AFX62711.1 AFYH01268658 AFYH01268659 JX656768 AFX62708.1 JX656770 AFX62710.1 JX656754 AFX62694.1 JX656774 AFX62714.1 JX656777 AFX62717.1 AMQN01007694 AMQN01007695 KB301197 ELU05817.1 JX656758 AFX62698.1 JX656767 AFX62707.1 JX656757 AFX62697.1 JX656769 AFX62709.1 JX656761 AFX62701.1 GBBM01003377 JAC32041.1 NEVH01006722 PNF37087.1 GFAC01001823 JAT97365.1 JX656753 AFX62693.1 JX656772 AFX62712.1 GAKP01019289 JAC39663.1 HADY01018577 SBP57062.1 CP001330 ACO66586.1 MF288057 AWH61374.1 JX656755 AFX62695.1 RQTK01000817 RUS74590.1 JH431789 MF288058 AWH61375.1 PBBL01000049 MBH24570.1 PZQS01000001 PVD39343.1 HAAD01002595 CDG68827.1 LT674435 SIP62991.1 GEDC01009914 JAS27384.1 NNAY01000206 OXU30037.1 DS469809 EDO32735.1 GEDV01003978 JAP84579.1 RCHS01001937 RMX50570.1 GFAA01002021 JAU01414.1 AGTO01020911 AMQM01003827 KB096325 ESO05597.1 JX656930 AFX62870.1 MBFU01000045 PWA02908.1 MBFT01000090 PVU98069.1 MF288060 AWH61377.1 MF288059 AWH61376.1 NEDP02002422 OWF50824.1 BFAA01000455 GCB72342.1 GFPF01004417 MAA15563.1 QUSF01011075 RLV62105.1 JX656937 AFX62877.1
KPI95442.1 KZ150012 PZC75052.1 JTDY01000843 KOB75693.1 ODYU01008518 SOQ52189.1 PGGS01000036 PNH11184.1 GECZ01008801 JAS60968.1 KK852464 KDR23312.1 GL378409 EFJ40702.1 GEBQ01030751 JAT09226.1 GECU01036892 JAS70814.1 CM008974 PNW74265.1 DS496125 GAMC01015305 GAMC01015303 JAB91252.1 JX656776 AFX62716.1 CH480831 EDW45865.1 JX656766 AFX62706.1 JX656764 JX656765 AFX62704.1 LT674447 SIP63003.1 AM294441 CM000361 CM002910 CAL26384.1 EDX04750.1 KMY89848.1 JX656759 AFX62699.1 JX656760 JX656762 AFX62702.1 GBXI01017267 JAC97024.1 JX656756 AFX62696.1 JX656763 JX656773 JX656775 AFX62713.1 JX656771 AFX62711.1 AFYH01268658 AFYH01268659 JX656768 AFX62708.1 JX656770 AFX62710.1 JX656754 AFX62694.1 JX656774 AFX62714.1 JX656777 AFX62717.1 AMQN01007694 AMQN01007695 KB301197 ELU05817.1 JX656758 AFX62698.1 JX656767 AFX62707.1 JX656757 AFX62697.1 JX656769 AFX62709.1 JX656761 AFX62701.1 GBBM01003377 JAC32041.1 NEVH01006722 PNF37087.1 GFAC01001823 JAT97365.1 JX656753 AFX62693.1 JX656772 AFX62712.1 GAKP01019289 JAC39663.1 HADY01018577 SBP57062.1 CP001330 ACO66586.1 MF288057 AWH61374.1 JX656755 AFX62695.1 RQTK01000817 RUS74590.1 JH431789 MF288058 AWH61375.1 PBBL01000049 MBH24570.1 PZQS01000001 PVD39343.1 HAAD01002595 CDG68827.1 LT674435 SIP62991.1 GEDC01009914 JAS27384.1 NNAY01000206 OXU30037.1 DS469809 EDO32735.1 GEDV01003978 JAP84579.1 RCHS01001937 RMX50570.1 GFAA01002021 JAU01414.1 AGTO01020911 AMQM01003827 KB096325 ESO05597.1 JX656930 AFX62870.1 MBFU01000045 PWA02908.1 MBFT01000090 PVU98069.1 MF288060 AWH61377.1 MF288059 AWH61376.1 NEDP02002422 OWF50824.1 BFAA01000455 GCB72342.1 GFPF01004417 MAA15563.1 QUSF01011075 RLV62105.1 JX656937 AFX62877.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000037510
UP000027135
+ More
UP000001058 UP000095301 UP000006906 UP000095300 UP000001292 UP000000304 UP000008672 UP000014760 UP000235965 UP000002009 UP000271974 UP000245119 UP000265040 UP000085678 UP000002280 UP000261420 UP000261360 UP000261600 UP000215335 UP000001593 UP000087266 UP000261400 UP000275408 UP000016665 UP000261640 UP000076420 UP000265140 UP000015101 UP000092443 UP000245591 UP000245699 UP000265200 UP000001038 UP000265180 UP000242188 UP000257160 UP000192221 UP000288216 UP000276834 UP000007635
UP000001058 UP000095301 UP000006906 UP000095300 UP000001292 UP000000304 UP000008672 UP000014760 UP000235965 UP000002009 UP000271974 UP000245119 UP000265040 UP000085678 UP000002280 UP000261420 UP000261360 UP000261600 UP000215335 UP000001593 UP000087266 UP000261400 UP000275408 UP000016665 UP000261640 UP000076420 UP000265140 UP000015101 UP000092443 UP000245591 UP000245699 UP000265200 UP000001038 UP000265180 UP000242188 UP000257160 UP000192221 UP000288216 UP000276834 UP000007635
PRIDE
Pfam
PF13091 PLDc_2
ProteinModelPortal
H9JEH1
A0A194QM63
A0A2A4K4V7
A0A194PQ50
A0A2W1BJ03
A0A0L7LJI0
+ More
A0A2H1WGJ8 A0A2J8AF98 A0A1B6GEV7 A0A067RHN9 D8UHV0 A0A1I8MWS3 A0A1B6KCR5 A0A1B6H873 A0A2K3D149 A8IW99 A0A1I8PG51 W8AYK0 K7WRZ7 B4IE38 K7WRZ0 K7WPV2 A0A1W1ELN7 A0AP53 K7WPV0 K7WKG9 A0A0A1WEX7 K7WRY1 K7XHQ2 K7WRZ4 H2ZTC7 K7XHQ0 K7X4V0 K7WPU5 K7WPV8 K7WKI0 R7UPE4 K7XHP5 K7WKH3 K7WKG6 K7WPV6 K7WRY6 A0A023GD06 A0A2J7R8C9 A0A1E1XDP6 K7XHP1 K7WKH6 A0A034VDP1 A0A1A8APG1 C1EET0 A0A2S1PS15 K7X4U0 A0A3S1B8F8 T1JLA2 A0A2S1PRW4 A0A2E5HM73 A0A2T7Q0Z7 T2MA50 A0A1W1ELM3 A0A3Q1H821 A0A1S3K043 F6TQM8 A0A3B4VN71 A0A3B4YFF5 A0A3Q3KC05 A0A1B6DNY1 A0A232FI96 A7SU88 A0A1S3Q697 A0A3B5ARB5 A0A1S3Q608 A0A1S3Q907 A0A131YZG8 A0A3M6UA87 A0A1E1XQA7 U3KH83 A0A3Q3RKB0 A0A2C9K659 A0A3P8Z8K9 T1EF80 A0A3P9ANA1 A0A1A9X7N3 K7X570 A0A2U1JD42 A0A2T9Z0I0 A0A2S1PRV2 A0A3P9HJQ8 H2LEV7 A0A3P9M3E3 A0A2S1PRU9 A0A210QQ12 A0A3Q1CKK2 A0A1W4VAK5 A0A401PGT5 A0A224YMV2 A0A3L8Q451 G3Q753 A0A3P8Z8M3 K7WKW4
A0A2H1WGJ8 A0A2J8AF98 A0A1B6GEV7 A0A067RHN9 D8UHV0 A0A1I8MWS3 A0A1B6KCR5 A0A1B6H873 A0A2K3D149 A8IW99 A0A1I8PG51 W8AYK0 K7WRZ7 B4IE38 K7WRZ0 K7WPV2 A0A1W1ELN7 A0AP53 K7WPV0 K7WKG9 A0A0A1WEX7 K7WRY1 K7XHQ2 K7WRZ4 H2ZTC7 K7XHQ0 K7X4V0 K7WPU5 K7WPV8 K7WKI0 R7UPE4 K7XHP5 K7WKH3 K7WKG6 K7WPV6 K7WRY6 A0A023GD06 A0A2J7R8C9 A0A1E1XDP6 K7XHP1 K7WKH6 A0A034VDP1 A0A1A8APG1 C1EET0 A0A2S1PS15 K7X4U0 A0A3S1B8F8 T1JLA2 A0A2S1PRW4 A0A2E5HM73 A0A2T7Q0Z7 T2MA50 A0A1W1ELM3 A0A3Q1H821 A0A1S3K043 F6TQM8 A0A3B4VN71 A0A3B4YFF5 A0A3Q3KC05 A0A1B6DNY1 A0A232FI96 A7SU88 A0A1S3Q697 A0A3B5ARB5 A0A1S3Q608 A0A1S3Q907 A0A131YZG8 A0A3M6UA87 A0A1E1XQA7 U3KH83 A0A3Q3RKB0 A0A2C9K659 A0A3P8Z8K9 T1EF80 A0A3P9ANA1 A0A1A9X7N3 K7X570 A0A2U1JD42 A0A2T9Z0I0 A0A2S1PRV2 A0A3P9HJQ8 H2LEV7 A0A3P9M3E3 A0A2S1PRU9 A0A210QQ12 A0A3Q1CKK2 A0A1W4VAK5 A0A401PGT5 A0A224YMV2 A0A3L8Q451 G3Q753 A0A3P8Z8M3 K7WKW4
PDB
4GEL
E-value=5.40401e-25,
Score=277
Ontologies
GO
GO:0003824
GO:0016021
GO:0016787
GO:0046872
GO:0004519
GO:0034587
GO:0051321
GO:0005741
GO:0016042
GO:0042802
GO:0031047
GO:0030717
GO:0016891
GO:0035755
GO:0043186
GO:0005739
GO:0042803
GO:0010636
GO:0030719
GO:0043046
GO:0007286
GO:0008053
GO:0006508
GO:0008061
GO:0007219
GO:0007275
GO:0050793
GO:0003676
GO:0016876
GO:0043039
Topology
Subcellular location
Mitochondrion outer membrane
Length:
142
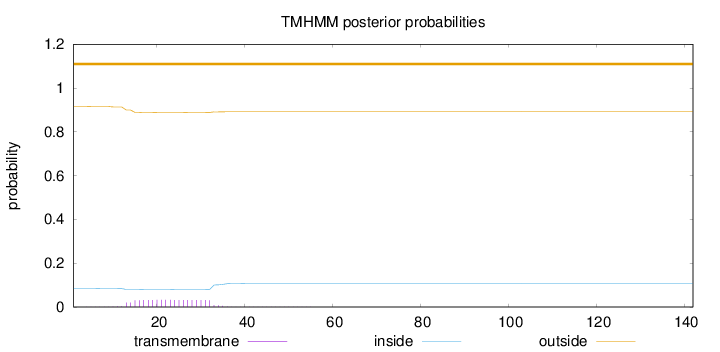
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.649019999999999
Exp number, first 60 AAs:
0.64705
Total prob of N-in:
0.08439
outside
1 - 142
Population Genetic Test Statistics
Pi
109.386188
Theta
120.733648
Tajima's D
-0.294307
CLR
0.095667
CSRT
0.290585470726464
Interpretation
Uncertain
