Gene
KWMTBOMO08716 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007919
Annotation
PREDICTED:_estradiol_17-beta-dehydrogenase_8-like_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 1.656
Sequence
CDS
ATGGTGTCGGGTTTAGTAGTAGGTCGCCTTGCTCTCGTTACTGGTGCCGGTTCTGGGATTGGTCGTGCAGCGTGCCAAGTTTTATCAAAAGAGGGAGCCACTGTGATAGCCGCAGACAGGAAGTATGAAGCTGCTGTACAAACTATAAAAACTCATACAGCATTAGCACCTGGTACTAATGGTGGTGAGGAGCATAGTGCTGTGGAATTAGACGTTTCCAACAGTAAAGCAGTCCAAGACCTGGTACAAGATACACTGAAGCAATATAAAAAGCCTCCTTGCATCATTGTCAACTCTGCCGGCATTACCAGGGATAATTGGATGCTGAAACTCTCAGAGGAAGACTATGATAGTGTGCTTAATGTAAACTTGAAGGGTACATTCTTAGTGATGCAGGCATTTGCAAAAGCGATGGCCGAGTCGTCAGTGACGGGTTCTATAATAAATATCTCTAGCATTGTTGGAAAATACGGAAACATGGGGCAGACAAACTATGCGAGTAGTAAGGCTGGAGTGGTCGCGATGACTCAATCCGCAGCCAAGGAGTTAGGAAAATTCAACATAAGGGTGAACGCAATCTTACCTGGATTCATTGACACACCGATAATAAAAACAGTCCCTGACAAAGTCAAGGAACAGATTGTGAAATTAGTCCCACTAGGACGACTCGGTAATCCTGCAGAAGTTGCAGAGGTAATCGCCTTCTTAAGTTCCGACAAGAGCTCTTTCATCACTGGTGCTGCTATTGACGTCAGTGGAGGGTATTAA
Protein
MVSGLVVGRLALVTGAGSGIGRAACQVLSKEGATVIAADRKYEAAVQTIKTHTALAPGTNGGEEHSAVELDVSNSKAVQDLVQDTLKQYKKPPCIIVNSAGITRDNWMLKLSEEDYDSVLNVNLKGTFLVMQAFAKAMAESSVTGSIINISSIVGKYGNMGQTNYASSKAGVVAMTQSAAKELGKFNIRVNAILPGFIDTPIIKTVPDKVKEQIVKLVPLGRLGNPAEVAEVIAFLSSDKSSFITGAAIDVSGGY
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JEH2
A0A212F641
A0A2H1WGJ6
A0A3S2LT13
I4DL53
A0A194QNS9
+ More
A0A2W1BL92 A0A194PWD0 A0A194QSW4 A0A2A4K4U4 A0A1L8DIW5 B4MCS4 B3MWD6 A0A1B0CE30 B4JJG0 A0A034WJ56 A0A1Y1MUQ4 A0A0K8U1E6 A0A0M4EMA7 D6X4G4 C0MM13 Q9W4U2 A0A0A1X145 A0A0B4J2T3 A0A195D3T0 A0A336M7G9 W8BRS7 A0A1I8Q7M2 A0A182GXJ0 A0A3B0K2R8 B4NDX7 B0W411 A0A1W4VRX8 A0A1I8Q7Q6 A0A182IW82 T1PGG1 B3NTZ1 A0A1I8N8R6 Q29HP4 B4L898 B4Q0U0 A0A232EGH7 A0A182Y8C7 B4R3V9 A0A023EN08 A0A1A9WL56 F4WKF5 A0A182XCE5 A0A182P1P4 A0A0L0C162 B4F625 A0A182L644 Q7Q1U0 A0A182ICQ7 A0A1Y1MN79 A0A182VTU3 A0A182U884 A0A182RGN2 A0A0K8WKQ7 B4IA21 A0A182MT92 A0A195DI61 A0A2A3EHF5 A0A084W1Y8 A0A2C9M9K3 A0A1I8JSL7 B4F631 A0A0M9A8F6 A0A182K4D5 A0A182SDD6 Q561S1 A0A182QC50 A0A2M4BY11 U5EVE1 W5J9L5 A0A1J1IMH0 J9I050 Q1LWX6 Q16F48 A0A2R7W0D0 V5GJ58 A0A2M3ZDI3 A0A195EQN1 A0A0L8GCE7 A0A2B4RVX5 A0A182NB02 E2B9T1 Q8AW16 A0A1S3H855 A7RXH2 A0A336K5N9 A0A0T6AYX8 A0A1S3JCP3 A0A3M6U1E4 A0A1B0G0Y9 A0A2M4AVP4 A0A2J7PQF3 A0A310SLF4 D3TL56 A0A158NQM7
A0A2W1BL92 A0A194PWD0 A0A194QSW4 A0A2A4K4U4 A0A1L8DIW5 B4MCS4 B3MWD6 A0A1B0CE30 B4JJG0 A0A034WJ56 A0A1Y1MUQ4 A0A0K8U1E6 A0A0M4EMA7 D6X4G4 C0MM13 Q9W4U2 A0A0A1X145 A0A0B4J2T3 A0A195D3T0 A0A336M7G9 W8BRS7 A0A1I8Q7M2 A0A182GXJ0 A0A3B0K2R8 B4NDX7 B0W411 A0A1W4VRX8 A0A1I8Q7Q6 A0A182IW82 T1PGG1 B3NTZ1 A0A1I8N8R6 Q29HP4 B4L898 B4Q0U0 A0A232EGH7 A0A182Y8C7 B4R3V9 A0A023EN08 A0A1A9WL56 F4WKF5 A0A182XCE5 A0A182P1P4 A0A0L0C162 B4F625 A0A182L644 Q7Q1U0 A0A182ICQ7 A0A1Y1MN79 A0A182VTU3 A0A182U884 A0A182RGN2 A0A0K8WKQ7 B4IA21 A0A182MT92 A0A195DI61 A0A2A3EHF5 A0A084W1Y8 A0A2C9M9K3 A0A1I8JSL7 B4F631 A0A0M9A8F6 A0A182K4D5 A0A182SDD6 Q561S1 A0A182QC50 A0A2M4BY11 U5EVE1 W5J9L5 A0A1J1IMH0 J9I050 Q1LWX6 Q16F48 A0A2R7W0D0 V5GJ58 A0A2M3ZDI3 A0A195EQN1 A0A0L8GCE7 A0A2B4RVX5 A0A182NB02 E2B9T1 Q8AW16 A0A1S3H855 A7RXH2 A0A336K5N9 A0A0T6AYX8 A0A1S3JCP3 A0A3M6U1E4 A0A1B0G0Y9 A0A2M4AVP4 A0A2J7PQF3 A0A310SLF4 D3TL56 A0A158NQM7
Pubmed
19121390
22118469
22651552
26354079
28756777
17994087
+ More
25348373 28004739 18362917 19820115 19126864 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 20075255 24495485 26483478 25315136 15632085 17550304 28648823 25244985 24945155 21719571 26108605 18477586 20966253 12364791 14747013 17210077 24438588 15562597 20920257 23761445 17510324 23594743 20798317 17615350 26383154 30382153 20353571 21347285
25348373 28004739 18362917 19820115 19126864 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 20075255 24495485 26483478 25315136 15632085 17550304 28648823 25244985 24945155 21719571 26108605 18477586 20966253 12364791 14747013 17210077 24438588 15562597 20920257 23761445 17510324 23594743 20798317 17615350 26383154 30382153 20353571 21347285
EMBL
BABH01003078
BABH01003079
AGBW02010081
OWR49198.1
ODYU01008518
SOQ52190.1
+ More
RSAL01000014 RVE53231.1 AK402021 BAM18643.1 KQ461195 KPJ06630.1 KZ150012 PZC75051.1 KQ459596 KPI95440.1 KPJ06631.1 NWSH01000128 PCG79275.1 GFDF01007702 JAV06382.1 CH940660 EDW57996.1 CH902625 EDV34921.2 AJWK01008531 CH916370 EDV99712.1 GAKP01004792 JAC54160.1 GEZM01026257 JAV87087.1 GDHF01031993 JAI20321.1 CP012528 ALC48821.1 KQ971380 EEZ97555.1 FM246385 CAR94311.1 AE014298 AY113563 AAF45844.2 AAM29568.1 AGB95045.1 GBXI01009505 JAD04787.1 AAZX01000353 KQ976885 KYN07511.1 UFQS01001153 UFQT01000663 UFQT01000664 UFQT01001153 SSX09275.1 SSX26264.1 SSX26265.1 GAMC01006962 JAB99593.1 JXUM01020458 JXUM01034336 JXUM01034337 KQ561020 KQ560572 KXJ80008.1 KXJ81856.1 OUUW01000015 SPP88524.1 CH964239 EDW81946.1 DS231834 EDS32604.1 KA647892 AFP62521.1 CH954180 EDV45699.1 CH379064 EAL31714.4 CH933814 EDW05673.1 CM000162 EDX01307.1 NNAY01004762 OXU17453.1 CM000366 EDX16971.1 GAPW01003338 JAC10260.1 GL888199 EGI65293.1 JRES01001043 KNC26001.1 AM999261 AM999262 AM999263 AM999264 AM999265 AM999266 AM999268 FM246374 FM246375 FM246376 FM246377 FM246378 FM246379 FM246380 FM246381 FM246382 FM246383 FM246384 CAQ53623.1 CAQ53624.1 CAQ53625.1 CAQ53626.1 CAQ53627.1 CAQ53628.1 CAQ53630.1 CAR94300.1 CAR94301.1 CAR94302.1 CAR94303.1 CAR94304.1 CAR94305.1 CAR94306.1 CAR94307.1 CAR94308.1 CAR94309.1 CAR94310.1 AAAB01008980 EAA13862.3 APCN01005749 GEZM01026255 JAV87089.1 GDHF01000632 JAI51682.1 CH480825 EDW44052.1 AXCM01006310 KQ980824 KYN12522.1 KZ288256 PBC30606.1 ATLV01019471 KE525272 KFB44232.1 AM999267 CAQ53629.1 KQ435712 KOX79360.1 BC093373 AAH93373.1 AXCN02000649 GGFJ01008753 MBW57894.1 GANO01001899 JAB57972.1 ADMH02001879 ETN60691.1 CVRI01000054 CRL00756.1 CH478512 EJY58115.1 FP085392 BX510994 CAK04961.1 EAT32860.1 KK854215 PTY13203.1 GALX01004362 JAB64104.1 GGFM01005800 MBW26551.1 KQ982021 KYN30585.1 KQ422595 KOF74613.1 LSMT01000315 PFX20487.1 GL446605 EFN87518.1 AL672176 CAD54662.1 DS469550 EDO43775.1 UFQS01000135 UFQT01000135 SSX00286.1 SSX20666.1 LJIG01022498 KRT80258.1 RCHS01002437 RMX47344.1 CCAG010003430 GGFK01011520 MBW44841.1 NEVH01022638 PNF18567.1 KQ760132 OAD61804.1 EZ422156 ADD18351.1 ADTU01023414
RSAL01000014 RVE53231.1 AK402021 BAM18643.1 KQ461195 KPJ06630.1 KZ150012 PZC75051.1 KQ459596 KPI95440.1 KPJ06631.1 NWSH01000128 PCG79275.1 GFDF01007702 JAV06382.1 CH940660 EDW57996.1 CH902625 EDV34921.2 AJWK01008531 CH916370 EDV99712.1 GAKP01004792 JAC54160.1 GEZM01026257 JAV87087.1 GDHF01031993 JAI20321.1 CP012528 ALC48821.1 KQ971380 EEZ97555.1 FM246385 CAR94311.1 AE014298 AY113563 AAF45844.2 AAM29568.1 AGB95045.1 GBXI01009505 JAD04787.1 AAZX01000353 KQ976885 KYN07511.1 UFQS01001153 UFQT01000663 UFQT01000664 UFQT01001153 SSX09275.1 SSX26264.1 SSX26265.1 GAMC01006962 JAB99593.1 JXUM01020458 JXUM01034336 JXUM01034337 KQ561020 KQ560572 KXJ80008.1 KXJ81856.1 OUUW01000015 SPP88524.1 CH964239 EDW81946.1 DS231834 EDS32604.1 KA647892 AFP62521.1 CH954180 EDV45699.1 CH379064 EAL31714.4 CH933814 EDW05673.1 CM000162 EDX01307.1 NNAY01004762 OXU17453.1 CM000366 EDX16971.1 GAPW01003338 JAC10260.1 GL888199 EGI65293.1 JRES01001043 KNC26001.1 AM999261 AM999262 AM999263 AM999264 AM999265 AM999266 AM999268 FM246374 FM246375 FM246376 FM246377 FM246378 FM246379 FM246380 FM246381 FM246382 FM246383 FM246384 CAQ53623.1 CAQ53624.1 CAQ53625.1 CAQ53626.1 CAQ53627.1 CAQ53628.1 CAQ53630.1 CAR94300.1 CAR94301.1 CAR94302.1 CAR94303.1 CAR94304.1 CAR94305.1 CAR94306.1 CAR94307.1 CAR94308.1 CAR94309.1 CAR94310.1 AAAB01008980 EAA13862.3 APCN01005749 GEZM01026255 JAV87089.1 GDHF01000632 JAI51682.1 CH480825 EDW44052.1 AXCM01006310 KQ980824 KYN12522.1 KZ288256 PBC30606.1 ATLV01019471 KE525272 KFB44232.1 AM999267 CAQ53629.1 KQ435712 KOX79360.1 BC093373 AAH93373.1 AXCN02000649 GGFJ01008753 MBW57894.1 GANO01001899 JAB57972.1 ADMH02001879 ETN60691.1 CVRI01000054 CRL00756.1 CH478512 EJY58115.1 FP085392 BX510994 CAK04961.1 EAT32860.1 KK854215 PTY13203.1 GALX01004362 JAB64104.1 GGFM01005800 MBW26551.1 KQ982021 KYN30585.1 KQ422595 KOF74613.1 LSMT01000315 PFX20487.1 GL446605 EFN87518.1 AL672176 CAD54662.1 DS469550 EDO43775.1 UFQS01000135 UFQT01000135 SSX00286.1 SSX20666.1 LJIG01022498 KRT80258.1 RCHS01002437 RMX47344.1 CCAG010003430 GGFK01011520 MBW44841.1 NEVH01022638 PNF18567.1 KQ760132 OAD61804.1 EZ422156 ADD18351.1 ADTU01023414
Proteomes
UP000005204
UP000007151
UP000283053
UP000053240
UP000053268
UP000218220
+ More
UP000008792 UP000007801 UP000092461 UP000001070 UP000092553 UP000007266 UP000000803 UP000002358 UP000078542 UP000095300 UP000069940 UP000249989 UP000268350 UP000007798 UP000002320 UP000192221 UP000075880 UP000008711 UP000095301 UP000001819 UP000009192 UP000002282 UP000215335 UP000076408 UP000000304 UP000091820 UP000007755 UP000076407 UP000075885 UP000037069 UP000075882 UP000007062 UP000075840 UP000075920 UP000075902 UP000075900 UP000001292 UP000075883 UP000078492 UP000242457 UP000030765 UP000076420 UP000069272 UP000053105 UP000075881 UP000075901 UP000075886 UP000000673 UP000183832 UP000008820 UP000000437 UP000078541 UP000053454 UP000225706 UP000075884 UP000008237 UP000085678 UP000001593 UP000275408 UP000092444 UP000235965 UP000005205
UP000008792 UP000007801 UP000092461 UP000001070 UP000092553 UP000007266 UP000000803 UP000002358 UP000078542 UP000095300 UP000069940 UP000249989 UP000268350 UP000007798 UP000002320 UP000192221 UP000075880 UP000008711 UP000095301 UP000001819 UP000009192 UP000002282 UP000215335 UP000076408 UP000000304 UP000091820 UP000007755 UP000076407 UP000075885 UP000037069 UP000075882 UP000007062 UP000075840 UP000075920 UP000075902 UP000075900 UP000001292 UP000075883 UP000078492 UP000242457 UP000030765 UP000076420 UP000069272 UP000053105 UP000075881 UP000075901 UP000075886 UP000000673 UP000183832 UP000008820 UP000000437 UP000078541 UP000053454 UP000225706 UP000075884 UP000008237 UP000085678 UP000001593 UP000275408 UP000092444 UP000235965 UP000005205
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JEH2
A0A212F641
A0A2H1WGJ6
A0A3S2LT13
I4DL53
A0A194QNS9
+ More
A0A2W1BL92 A0A194PWD0 A0A194QSW4 A0A2A4K4U4 A0A1L8DIW5 B4MCS4 B3MWD6 A0A1B0CE30 B4JJG0 A0A034WJ56 A0A1Y1MUQ4 A0A0K8U1E6 A0A0M4EMA7 D6X4G4 C0MM13 Q9W4U2 A0A0A1X145 A0A0B4J2T3 A0A195D3T0 A0A336M7G9 W8BRS7 A0A1I8Q7M2 A0A182GXJ0 A0A3B0K2R8 B4NDX7 B0W411 A0A1W4VRX8 A0A1I8Q7Q6 A0A182IW82 T1PGG1 B3NTZ1 A0A1I8N8R6 Q29HP4 B4L898 B4Q0U0 A0A232EGH7 A0A182Y8C7 B4R3V9 A0A023EN08 A0A1A9WL56 F4WKF5 A0A182XCE5 A0A182P1P4 A0A0L0C162 B4F625 A0A182L644 Q7Q1U0 A0A182ICQ7 A0A1Y1MN79 A0A182VTU3 A0A182U884 A0A182RGN2 A0A0K8WKQ7 B4IA21 A0A182MT92 A0A195DI61 A0A2A3EHF5 A0A084W1Y8 A0A2C9M9K3 A0A1I8JSL7 B4F631 A0A0M9A8F6 A0A182K4D5 A0A182SDD6 Q561S1 A0A182QC50 A0A2M4BY11 U5EVE1 W5J9L5 A0A1J1IMH0 J9I050 Q1LWX6 Q16F48 A0A2R7W0D0 V5GJ58 A0A2M3ZDI3 A0A195EQN1 A0A0L8GCE7 A0A2B4RVX5 A0A182NB02 E2B9T1 Q8AW16 A0A1S3H855 A7RXH2 A0A336K5N9 A0A0T6AYX8 A0A1S3JCP3 A0A3M6U1E4 A0A1B0G0Y9 A0A2M4AVP4 A0A2J7PQF3 A0A310SLF4 D3TL56 A0A158NQM7
A0A2W1BL92 A0A194PWD0 A0A194QSW4 A0A2A4K4U4 A0A1L8DIW5 B4MCS4 B3MWD6 A0A1B0CE30 B4JJG0 A0A034WJ56 A0A1Y1MUQ4 A0A0K8U1E6 A0A0M4EMA7 D6X4G4 C0MM13 Q9W4U2 A0A0A1X145 A0A0B4J2T3 A0A195D3T0 A0A336M7G9 W8BRS7 A0A1I8Q7M2 A0A182GXJ0 A0A3B0K2R8 B4NDX7 B0W411 A0A1W4VRX8 A0A1I8Q7Q6 A0A182IW82 T1PGG1 B3NTZ1 A0A1I8N8R6 Q29HP4 B4L898 B4Q0U0 A0A232EGH7 A0A182Y8C7 B4R3V9 A0A023EN08 A0A1A9WL56 F4WKF5 A0A182XCE5 A0A182P1P4 A0A0L0C162 B4F625 A0A182L644 Q7Q1U0 A0A182ICQ7 A0A1Y1MN79 A0A182VTU3 A0A182U884 A0A182RGN2 A0A0K8WKQ7 B4IA21 A0A182MT92 A0A195DI61 A0A2A3EHF5 A0A084W1Y8 A0A2C9M9K3 A0A1I8JSL7 B4F631 A0A0M9A8F6 A0A182K4D5 A0A182SDD6 Q561S1 A0A182QC50 A0A2M4BY11 U5EVE1 W5J9L5 A0A1J1IMH0 J9I050 Q1LWX6 Q16F48 A0A2R7W0D0 V5GJ58 A0A2M3ZDI3 A0A195EQN1 A0A0L8GCE7 A0A2B4RVX5 A0A182NB02 E2B9T1 Q8AW16 A0A1S3H855 A7RXH2 A0A336K5N9 A0A0T6AYX8 A0A1S3JCP3 A0A3M6U1E4 A0A1B0G0Y9 A0A2M4AVP4 A0A2J7PQF3 A0A310SLF4 D3TL56 A0A158NQM7
PDB
2PD6
E-value=9.32658e-64,
Score=615
Ontologies
PATHWAY
GO
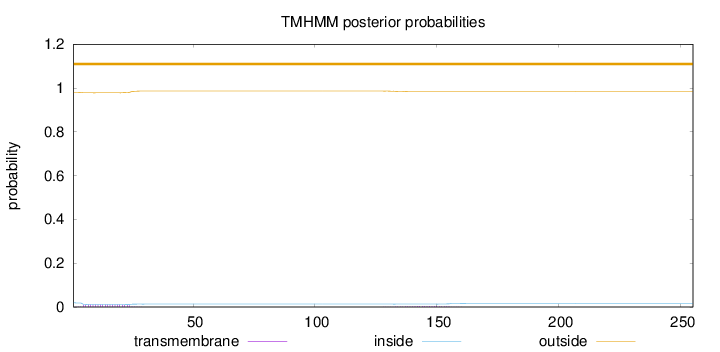
Topology
Length:
255
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29637
Exp number, first 60 AAs:
0.21294
Total prob of N-in:
0.01988
outside
1 - 255
Population Genetic Test Statistics
Pi
274.829566
Theta
180.531339
Tajima's D
1.287957
CLR
12.273281
CSRT
0.733063346832658
Interpretation
Uncertain
