Gene
KWMTBOMO08714
Pre Gene Modal
BGIBMGA007668
Annotation
PREDICTED:_chymotrypsin-2-like_[Amyelois_transitella]
Full name
Chymotrypsin-2
Alternative Name
AnChym2
Location in the cell
Extracellular Reliability : 2.686
Sequence
CDS
ATGTCACAGGATTCACCACGAGTGGTAGGCGGTGTGACTGTACCAGATGGTCGTATCCCTTATCAGGCGTCTCTGCGAACACTCTTCAATTCTCACTTTTGTGGCGGAAGTATCCTTAACGAGAAATGGGTCTTGACTGCCGCGCACTGTACTGTTGGCCAATCACATTTGACAATGCAAGTTGTGGTTGGAACAAACAATCTCCACATAGGCGGCGAGCACCACTCCGTCGAGTCGATCATCATACATAGTGACTACAACAGCATGGCCATCAGCAATGACGTCAGCCTGGTGAAGGTATCTAATGACATCGTGTTCAATGATAACGTTCAGTCAATACAACTGCCTGACGATGACACGGACGTCGGAGCTGACGTCATGCTGAGTGGGTGGGGGAGACTTTCGTATCCAGGGGTATACCCCAATCACCTGCAAATGCTAAATCTAACATCTCTGAGTGTGGAGCTATGCCAGGATATCTTCGTTGGCATCAACCCTGTATTCTCGACTCAGATATGCGCCATGACCAAGGTTGGAGAAGGAGCATGCCACGGCGATTCTGGTGGTCCGTTGGTGGAAGGAAACACGGTGGTGGGCGTGGTGTCCTGGGGTATGCCCTGTGCTCGTGGCTACCCTGATGTATACTCAAGGGTCTTCGCATTTAAAGACTGGATATTGGAGAAAATGGAACAAAACTAA
Protein
MSQDSPRVVGGVTVPDGRIPYQASLRTLFNSHFCGGSILNEKWVLTAAHCTVGQSHLTMQVVVGTNNLHIGGEHHSVESIIIHSDYNSMAISNDVSLVKVSNDIVFNDNVQSIQLPDDDTDVGADVMLSGWGRLSYPGVYPNHLQMLNLTSLSVELCQDIFVGINPVFSTQICAMTKVGEGACHGDSGGPLVEGNTVVGVVSWGMPCARGYPDVYSRVFAFKDWILEKMEQN
Summary
Catalytic Activity
Preferential cleavage: Tyr-|-Xaa, Trp-|-Xaa, Phe-|-Xaa, Leu-|-Xaa.
Similarity
Belongs to the peptidase S1 family.
Keywords
Complete proteome
Digestion
Disulfide bond
Hydrolase
Protease
Reference proteome
Secreted
Serine protease
Signal
Zymogen
Feature
propeptide Activation peptide
chain Chymotrypsin-2
chain Chymotrypsin-2
Uniprot
H9JDS1
A0A2W1BJK4
A0A2A4K680
A0A3S2P5Y9
A0A194QMQ6
A0A194PRR2
+ More
A0A212F636 A0A212F633 A0A3S2TRK4 A0A2H1WU74 K7IRM7 A0A2A4JXA5 A0A2W1BDL5 A0A212ELH6 K7IMT5 A0A087ZQ85 A0A3S2NS83 K7IMT7 A0A2A3EKL4 K7IMT4 A0A0M9A2K3 A0A2H1WEZ5 K7IMT3 A0A0N1PJ78 A0A154P3U3 A0A195BZ04 A0A158P3L4 A0A158P3L3 A0A151IHB1 A0A0J7KPH6 A0A232EZ15 A0A194PZ53 E2ACM3 A0A026WVM2 A0A232EXJ7 D0R8Q9 H9JWP8 D6WK62 A0A1Y1MAP6 Q17IG9 A0A0L7R485 A0A151XG10 Q175P6 A0A232FHI8 D0R8R4 Q8T639 A0A023EMN4 A0A1Q3FSI0 A0A195F0A9 A0A1W7R7W0 W5JMT6 O97099 A0A151IT67 T1DGK1 A0A1Y1MKE8 Q9NGY6 A0A1J1HSC6 A0A336MC30 A0A1B0CGA7 A0A182JLT2 A0A1B0DFH7 A0A182UFJ4 B0WLB8 Q5TWQ5 A0A182KQG2 A0A2C9GS03 A0A182GGY4 A0A182VGU5 A0A1W7R806 A0A1B0GHG0 B0WAK4 A0A1B0DDL4 A0A1Q3FNV0 D0V550 A0A336M0L8 A0A1J1IR33 A0A1I8MI67 A0A0M5JCH1 A0A232FJD4 W5JVV6 O97097 A0A1S4FE82 A0A154P6Q3 K7J5P6 Q175L8 A0A182QXH3 O97100 Q16NM7 A0A0C9R4S6 Q9XY45 E9J8Q2 B0WHA0 A0A0U1Z1T2 A0A2C9GRR7 A0A182VP77 Q7QIZ2 Q17025 Q5TWE1 Q175P5 A0A182KLE4
A0A212F636 A0A212F633 A0A3S2TRK4 A0A2H1WU74 K7IRM7 A0A2A4JXA5 A0A2W1BDL5 A0A212ELH6 K7IMT5 A0A087ZQ85 A0A3S2NS83 K7IMT7 A0A2A3EKL4 K7IMT4 A0A0M9A2K3 A0A2H1WEZ5 K7IMT3 A0A0N1PJ78 A0A154P3U3 A0A195BZ04 A0A158P3L4 A0A158P3L3 A0A151IHB1 A0A0J7KPH6 A0A232EZ15 A0A194PZ53 E2ACM3 A0A026WVM2 A0A232EXJ7 D0R8Q9 H9JWP8 D6WK62 A0A1Y1MAP6 Q17IG9 A0A0L7R485 A0A151XG10 Q175P6 A0A232FHI8 D0R8R4 Q8T639 A0A023EMN4 A0A1Q3FSI0 A0A195F0A9 A0A1W7R7W0 W5JMT6 O97099 A0A151IT67 T1DGK1 A0A1Y1MKE8 Q9NGY6 A0A1J1HSC6 A0A336MC30 A0A1B0CGA7 A0A182JLT2 A0A1B0DFH7 A0A182UFJ4 B0WLB8 Q5TWQ5 A0A182KQG2 A0A2C9GS03 A0A182GGY4 A0A182VGU5 A0A1W7R806 A0A1B0GHG0 B0WAK4 A0A1B0DDL4 A0A1Q3FNV0 D0V550 A0A336M0L8 A0A1J1IR33 A0A1I8MI67 A0A0M5JCH1 A0A232FJD4 W5JVV6 O97097 A0A1S4FE82 A0A154P6Q3 K7J5P6 Q175L8 A0A182QXH3 O97100 Q16NM7 A0A0C9R4S6 Q9XY45 E9J8Q2 B0WHA0 A0A0U1Z1T2 A0A2C9GRR7 A0A182VP77 Q7QIZ2 Q17025 Q5TWE1 Q175P5 A0A182KLE4
EC Number
3.4.21.1
Pubmed
EMBL
BABH01003079
KZ150012
PZC75048.1
NWSH01000128
PCG79273.1
RSAL01000014
+ More
RVE53228.1 KQ461195 KPJ06629.1 KQ459596 KPI95439.1 AGBW02010081 OWR49200.1 OWR49201.1 RVE53229.1 ODYU01011093 SOQ56615.1 NWSH01000497 PCG76022.1 KZ150244 PZC71884.1 AGBW02014066 OWR42328.1 RSAL01000269 RVE43195.1 KZ288229 PBC31726.1 KQ435756 KOX75917.1 ODYU01007885 SOQ51044.1 KQ460599 KPJ13815.1 KQ434809 KZC06605.1 KQ976394 KYM93148.1 ADTU01008199 KQ977636 KYN01157.1 LBMM01004589 KMQ92263.1 NNAY01001573 OXU23549.1 KQ459583 KPI98612.1 GL438568 EFN68767.1 KK107109 QOIP01000008 EZA59134.1 RLU19794.1 NNAY01001736 OXU23082.1 FN547058 KQ971342 CBC01166.1 EFA04659.1 BABH01018411 EFA04622.1 GEZM01036400 GEZM01036398 GEZM01036395 GEZM01036393 GEZM01036392 GEZM01036391 JAV82733.1 CH477239 EAT46500.1 KQ414658 KOC65663.1 KQ982174 KYQ59316.1 CH477397 EAT41804.1 NNAY01000219 OXU29928.1 FN547063 CBC01177.1 AF487334 AAL93243.1 GAPW01003508 JAC10090.1 GFDL01004672 JAV30373.1 KQ981880 KYN33913.1 GEHC01000393 JAV47252.1 ADMH02001109 ETN64084.1 AF051780 AAD17493.1 KQ981039 KYN10142.1 GAMD01002746 JAA98844.1 GEZM01028907 JAV86199.1 AF237415 AAF43707.1 CVRI01000012 CRK89382.1 UFQT01000369 SSX23608.1 AJWK01010930 AJWK01010931 AJWK01010932 AJVK01005806 DS232962 DS231983 EDS27062.1 EDS30402.1 AAAB01008807 EAL41865.3 APCN01000169 JXUM01062824 KQ562217 KXJ76377.1 GEHC01000378 JAV47267.1 AJWK01004566 DS231873 EDS41558.1 AJVK01005414 GFDL01005774 JAV29271.1 GU018015 ACY24348.1 SSX23610.1 CVRI01000058 CRL02608.1 CP012526 ALC46481.1 NNAY01000131 OXU30693.1 ADMH02000310 ETN67024.1 AF051778 AAD17491.1 KQ434827 KZC07503.1 CH477398 EAT41784.1 AXCN02000582 AF051781 AAD17494.1 CH477816 EAT35957.1 EAT35958.1 GBYB01001836 JAG71603.1 AF053903 AAD21823.1 GL769036 EFZ10800.1 DS231933 EDS27558.1 KJ801816 AJO53282.1 EAA04161.3 Z18888 Z32645 EAL41979.1 EAT41805.1
RVE53228.1 KQ461195 KPJ06629.1 KQ459596 KPI95439.1 AGBW02010081 OWR49200.1 OWR49201.1 RVE53229.1 ODYU01011093 SOQ56615.1 NWSH01000497 PCG76022.1 KZ150244 PZC71884.1 AGBW02014066 OWR42328.1 RSAL01000269 RVE43195.1 KZ288229 PBC31726.1 KQ435756 KOX75917.1 ODYU01007885 SOQ51044.1 KQ460599 KPJ13815.1 KQ434809 KZC06605.1 KQ976394 KYM93148.1 ADTU01008199 KQ977636 KYN01157.1 LBMM01004589 KMQ92263.1 NNAY01001573 OXU23549.1 KQ459583 KPI98612.1 GL438568 EFN68767.1 KK107109 QOIP01000008 EZA59134.1 RLU19794.1 NNAY01001736 OXU23082.1 FN547058 KQ971342 CBC01166.1 EFA04659.1 BABH01018411 EFA04622.1 GEZM01036400 GEZM01036398 GEZM01036395 GEZM01036393 GEZM01036392 GEZM01036391 JAV82733.1 CH477239 EAT46500.1 KQ414658 KOC65663.1 KQ982174 KYQ59316.1 CH477397 EAT41804.1 NNAY01000219 OXU29928.1 FN547063 CBC01177.1 AF487334 AAL93243.1 GAPW01003508 JAC10090.1 GFDL01004672 JAV30373.1 KQ981880 KYN33913.1 GEHC01000393 JAV47252.1 ADMH02001109 ETN64084.1 AF051780 AAD17493.1 KQ981039 KYN10142.1 GAMD01002746 JAA98844.1 GEZM01028907 JAV86199.1 AF237415 AAF43707.1 CVRI01000012 CRK89382.1 UFQT01000369 SSX23608.1 AJWK01010930 AJWK01010931 AJWK01010932 AJVK01005806 DS232962 DS231983 EDS27062.1 EDS30402.1 AAAB01008807 EAL41865.3 APCN01000169 JXUM01062824 KQ562217 KXJ76377.1 GEHC01000378 JAV47267.1 AJWK01004566 DS231873 EDS41558.1 AJVK01005414 GFDL01005774 JAV29271.1 GU018015 ACY24348.1 SSX23610.1 CVRI01000058 CRL02608.1 CP012526 ALC46481.1 NNAY01000131 OXU30693.1 ADMH02000310 ETN67024.1 AF051778 AAD17491.1 KQ434827 KZC07503.1 CH477398 EAT41784.1 AXCN02000582 AF051781 AAD17494.1 CH477816 EAT35957.1 EAT35958.1 GBYB01001836 JAG71603.1 AF053903 AAD21823.1 GL769036 EFZ10800.1 DS231933 EDS27558.1 KJ801816 AJO53282.1 EAA04161.3 Z18888 Z32645 EAL41979.1 EAT41805.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000002358 UP000005203 UP000242457 UP000053105 UP000076502 UP000078540 UP000005205 UP000078542 UP000036403 UP000215335 UP000000311 UP000053097 UP000279307 UP000007266 UP000008820 UP000053825 UP000075809 UP000078541 UP000000673 UP000078492 UP000183832 UP000092461 UP000075880 UP000092462 UP000075902 UP000002320 UP000007062 UP000075882 UP000075840 UP000069940 UP000249989 UP000075903 UP000095301 UP000092553 UP000075886
UP000002358 UP000005203 UP000242457 UP000053105 UP000076502 UP000078540 UP000005205 UP000078542 UP000036403 UP000215335 UP000000311 UP000053097 UP000279307 UP000007266 UP000008820 UP000053825 UP000075809 UP000078541 UP000000673 UP000078492 UP000183832 UP000092461 UP000075880 UP000092462 UP000075902 UP000002320 UP000007062 UP000075882 UP000075840 UP000069940 UP000249989 UP000075903 UP000095301 UP000092553 UP000075886
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
H9JDS1
A0A2W1BJK4
A0A2A4K680
A0A3S2P5Y9
A0A194QMQ6
A0A194PRR2
+ More
A0A212F636 A0A212F633 A0A3S2TRK4 A0A2H1WU74 K7IRM7 A0A2A4JXA5 A0A2W1BDL5 A0A212ELH6 K7IMT5 A0A087ZQ85 A0A3S2NS83 K7IMT7 A0A2A3EKL4 K7IMT4 A0A0M9A2K3 A0A2H1WEZ5 K7IMT3 A0A0N1PJ78 A0A154P3U3 A0A195BZ04 A0A158P3L4 A0A158P3L3 A0A151IHB1 A0A0J7KPH6 A0A232EZ15 A0A194PZ53 E2ACM3 A0A026WVM2 A0A232EXJ7 D0R8Q9 H9JWP8 D6WK62 A0A1Y1MAP6 Q17IG9 A0A0L7R485 A0A151XG10 Q175P6 A0A232FHI8 D0R8R4 Q8T639 A0A023EMN4 A0A1Q3FSI0 A0A195F0A9 A0A1W7R7W0 W5JMT6 O97099 A0A151IT67 T1DGK1 A0A1Y1MKE8 Q9NGY6 A0A1J1HSC6 A0A336MC30 A0A1B0CGA7 A0A182JLT2 A0A1B0DFH7 A0A182UFJ4 B0WLB8 Q5TWQ5 A0A182KQG2 A0A2C9GS03 A0A182GGY4 A0A182VGU5 A0A1W7R806 A0A1B0GHG0 B0WAK4 A0A1B0DDL4 A0A1Q3FNV0 D0V550 A0A336M0L8 A0A1J1IR33 A0A1I8MI67 A0A0M5JCH1 A0A232FJD4 W5JVV6 O97097 A0A1S4FE82 A0A154P6Q3 K7J5P6 Q175L8 A0A182QXH3 O97100 Q16NM7 A0A0C9R4S6 Q9XY45 E9J8Q2 B0WHA0 A0A0U1Z1T2 A0A2C9GRR7 A0A182VP77 Q7QIZ2 Q17025 Q5TWE1 Q175P5 A0A182KLE4
A0A212F636 A0A212F633 A0A3S2TRK4 A0A2H1WU74 K7IRM7 A0A2A4JXA5 A0A2W1BDL5 A0A212ELH6 K7IMT5 A0A087ZQ85 A0A3S2NS83 K7IMT7 A0A2A3EKL4 K7IMT4 A0A0M9A2K3 A0A2H1WEZ5 K7IMT3 A0A0N1PJ78 A0A154P3U3 A0A195BZ04 A0A158P3L4 A0A158P3L3 A0A151IHB1 A0A0J7KPH6 A0A232EZ15 A0A194PZ53 E2ACM3 A0A026WVM2 A0A232EXJ7 D0R8Q9 H9JWP8 D6WK62 A0A1Y1MAP6 Q17IG9 A0A0L7R485 A0A151XG10 Q175P6 A0A232FHI8 D0R8R4 Q8T639 A0A023EMN4 A0A1Q3FSI0 A0A195F0A9 A0A1W7R7W0 W5JMT6 O97099 A0A151IT67 T1DGK1 A0A1Y1MKE8 Q9NGY6 A0A1J1HSC6 A0A336MC30 A0A1B0CGA7 A0A182JLT2 A0A1B0DFH7 A0A182UFJ4 B0WLB8 Q5TWQ5 A0A182KQG2 A0A2C9GS03 A0A182GGY4 A0A182VGU5 A0A1W7R806 A0A1B0GHG0 B0WAK4 A0A1B0DDL4 A0A1Q3FNV0 D0V550 A0A336M0L8 A0A1J1IR33 A0A1I8MI67 A0A0M5JCH1 A0A232FJD4 W5JVV6 O97097 A0A1S4FE82 A0A154P6Q3 K7J5P6 Q175L8 A0A182QXH3 O97100 Q16NM7 A0A0C9R4S6 Q9XY45 E9J8Q2 B0WHA0 A0A0U1Z1T2 A0A2C9GRR7 A0A182VP77 Q7QIZ2 Q17025 Q5TWE1 Q175P5 A0A182KLE4
PDB
1EQ9
E-value=5.08642e-45,
Score=453
Ontologies
GO
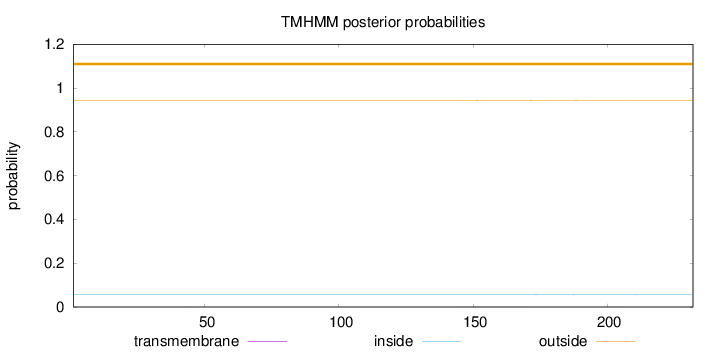
Topology
Subcellular location
Secreted
Length:
232
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01209
Exp number, first 60 AAs:
0.00317
Total prob of N-in:
0.05728
outside
1 - 232
Population Genetic Test Statistics
Pi
185.494567
Theta
152.31005
Tajima's D
0.575936
CLR
0.071011
CSRT
0.536523173841308
Interpretation
Uncertain
