Gene
KWMTBOMO08713
Annotation
unnamed_protein_product_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.348
Sequence
CDS
ATGGAGTTATCTGCTGCAAGTCTCTTAACACAGCTTATGATACGTGTCTCGACTCAATTGTTGTCCCATATTACCATAAAGCAACACATATGCTGGTCAGACAGTACCATTGTCCTAGCATGGCTAAACACACCTCCTCATAGACTTCAGGTCTTTGAAGCTAACAGGGTTGCCAAGATAACATCCAATCCAATCACCTCCACATGGAAACATGTTCCTACAAACCTGAACCCTGCAGACTGTGCGAGCAGGGGAATGTCTGCCCAGTCACTATCGGCTCATGATCTTTGGTGGTCTCCTAGCTGGCTGAAGGAGCCTCCTGACACCTGGCCCAAAATGCCTCCTGCTCTAGGCCACCATGCATTACCAGGTCTGAAGCCCAAGAAAGTACCAGCTCATATAGCAGTTCCTGACCTCGACCTTGACTTGCTTACTCGATTCAGTTCTCTCGACAAGCTCGTTGGAGTGACAGCTTGCATAAAACGCTTTATTTTCAATTGTAGACACAACTCAACTGATAGGCGCTCTGGTCCCTTGACAGTTGGAGAACGTAGAGATGCTCTGTTATTCTGGGTTCGCTCTGTTCAACACAATGAGTTTGCGGAAGATATTTACCGTCTTCAGGCAGGCAAGATTTGTACCGTTCGACTTCAGCGTCTTTCACCCTTGATGAAAGATGATCTCTTGAGAGTTGGTGGTCGCCTCACTCATGCGCCAATAAGATACGATGCTCAGCATCCTCTCGTTCTGCCCAGCTCTAGCCCACTAGTTGATCTCATCGTAGACCACTATCACAGAATAAACTGTCATCCTGGGGCTGACACATTACACGCTATCCTCCGTCAGCAGTTCTGGATACTCTCAGCACGTCGTGTCATCAGGCATCGCGTGTACAAATGCATTAGATGTTTCCGATATCGCGCTCAGGCCAGAGCTCCGTTGATGGCTGATTTGCCCGCAGACAGGGTCACACCCCAGCCACTCATCCAAGCTCTCTCTCAGCGTTTTTGGACTCTGTGGACCAAATCCTGTCTCCATACCCTCCAAACCCGCTCCAAATGGCTTTCCACATCCTCACCTCCTCATGTTGGAGAGCTCGTCCTCATCAAAGAGGACAATCAGCCCCCGCTGCAATGGAAACTTGGAAGGATTATCCGCACGTTACCCGGCAAGGATGGAGTGATCAGAGTCGTGGATTTGGACACGAAGCATGGAAGTCTGCGGAGACCTGTGTTCAAGTTAGCCAGGTTGCCGCTGGATTCAGCACATGAAGAAGTTCAGCCCCGGCCGCCCCAGGATTAG
Protein
MELSAASLLTQLMIRVSTQLLSHITIKQHICWSDSTIVLAWLNTPPHRLQVFEANRVAKITSNPITSTWKHVPTNLNPADCASRGMSAQSLSAHDLWWSPSWLKEPPDTWPKMPPALGHHALPGLKPKKVPAHIAVPDLDLDLLTRFSSLDKLVGVTACIKRFIFNCRHNSTDRRSGPLTVGERRDALLFWVRSVQHNEFAEDIYRLQAGKICTVRLQRLSPLMKDDLLRVGGRLTHAPIRYDAQHPLVLPSSSPLVDLIVDHYHRINCHPGADTLHAILRQQFWILSARRVIRHRVYKCIRCFRYRAQARAPLMADLPADRVTPQPLIQALSQRFWTLWTKSCLHTLQTRSKWLSTSSPPHVGELVLIKEDNQPPLQWKLGRIIRTLPGKDGVIRVVDLDTKHGSLRRPVFKLARLPLDSAHEEVQPRPPQD
Summary
Uniprot
Q9TYB5
O77042
D7EKN1
A0A1B6LDH0
A0A1B6LL14
A0A1B6GMH9
+ More
A0A1B6JE15 A0A1B6KZV2 A0A1B6MUZ9 A0A0A9XS77 J9M1J7 A0A2S2N9D0 A0A1S3DM66 A0A0A9YU87 V5GHM7 A0A0A9ZC74 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6MFR0 A0A0A9Z7S5 J9JVD6 A0A1S3DTZ4 A0A0A9Z7Z7 A0A1B6KG61 A0A146M0N4 A0A0A9X3X8 A0A0A9WP39 A0A0A9XUK1 A0A146KW37 X1XTX7 A0A146LZC3 A0A0A9XKM2 A0A0A9YNG0 A0A0K8SX96 A0A1S3DGE3 A0A0A9Z001 X1XMM8 A0A0A9VSF7 A0A0A9VYL4 A0A1S3DRF1 A0A1S3DNP1 J9KD60 A0A2G3AQ28 A0A3Q0JP94 X1XIA7 A0A2S2P963 V5GH76 A0A146LEU7 A0A2G3AQC0 X1WY95 A0A0A9WJX2 A0A2S2PM34 A0A1S3DM17 A0A2S2PED9 A0A0J7KBZ0 A0A1U8N896 A0A139W8E1 A0A1Y1LIG5 A0A2S2NCY5 A0A1Y1KVX6 A0A224XG39 A0A0J7KCR4 A0A023EYQ7 A0A023EYA4 A0A3L8DF94 A0A0A9YJ91 A0A0A9XX98 A0A0A9YQQ8 J9L9H5 A0A0A9YU99 A0A0L7KUL1 A0A0J7K7S0 A0A0A9WXF2 X1X412 A0A1S3DBK1 A0A1S4EIX5 A0A226CW75 A0A0J7JZS0 A0A0A9WW84 X1WSS7 A0A226DMG9 A0A226CWA5 A0A1S3DIZ2 A0A0A9VUR1 A0A226E226 S4NIN9 A0A3L8DEE4 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A224XGH3 X1WZY4 X1WYQ8 A0A146LBU6 A0A2S2R5S9 A0A0K8T5S0 A0A0A9WQV8 A0A0C9QLM0
A0A1B6JE15 A0A1B6KZV2 A0A1B6MUZ9 A0A0A9XS77 J9M1J7 A0A2S2N9D0 A0A1S3DM66 A0A0A9YU87 V5GHM7 A0A0A9ZC74 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6MFR0 A0A0A9Z7S5 J9JVD6 A0A1S3DTZ4 A0A0A9Z7Z7 A0A1B6KG61 A0A146M0N4 A0A0A9X3X8 A0A0A9WP39 A0A0A9XUK1 A0A146KW37 X1XTX7 A0A146LZC3 A0A0A9XKM2 A0A0A9YNG0 A0A0K8SX96 A0A1S3DGE3 A0A0A9Z001 X1XMM8 A0A0A9VSF7 A0A0A9VYL4 A0A1S3DRF1 A0A1S3DNP1 J9KD60 A0A2G3AQ28 A0A3Q0JP94 X1XIA7 A0A2S2P963 V5GH76 A0A146LEU7 A0A2G3AQC0 X1WY95 A0A0A9WJX2 A0A2S2PM34 A0A1S3DM17 A0A2S2PED9 A0A0J7KBZ0 A0A1U8N896 A0A139W8E1 A0A1Y1LIG5 A0A2S2NCY5 A0A1Y1KVX6 A0A224XG39 A0A0J7KCR4 A0A023EYQ7 A0A023EYA4 A0A3L8DF94 A0A0A9YJ91 A0A0A9XX98 A0A0A9YQQ8 J9L9H5 A0A0A9YU99 A0A0L7KUL1 A0A0J7K7S0 A0A0A9WXF2 X1X412 A0A1S3DBK1 A0A1S4EIX5 A0A226CW75 A0A0J7JZS0 A0A0A9WW84 X1WSS7 A0A226DMG9 A0A226CWA5 A0A1S3DIZ2 A0A0A9VUR1 A0A226E226 S4NIN9 A0A3L8DEE4 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A224XGH3 X1WZY4 X1WYQ8 A0A146LBU6 A0A2S2R5S9 A0A0K8T5S0 A0A0A9WQV8 A0A0C9QLM0
Pubmed
EMBL
AB012907
BAA33577.1
AB012906
BAA33576.1
DS497760
EFA13221.1
+ More
GEBQ01018232 JAT21745.1 GEBQ01015599 JAT24378.1 GECZ01006169 JAS63600.1 GECU01010337 JAS97369.1 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 GBHO01021103 GBHO01021101 JAG22501.1 JAG22503.1 ABLF02052963 GGMR01000737 MBY13356.1 GBHO01008418 GBHO01008416 JAG35186.1 JAG35188.1 GALX01004947 JAB63519.1 GBHO01004194 JAG39410.1 GEBQ01005223 JAT34754.1 GBHO01004196 JAG39408.1 ABLF02008594 GBHO01003578 JAG40026.1 GEBQ01029803 JAT10174.1 GDHC01006469 JAQ12160.1 GBHO01030122 GBHO01030121 GBHO01030120 GBHO01030116 JAG13482.1 JAG13483.1 JAG13484.1 JAG13488.1 GBHO01034065 GBHO01034061 JAG09539.1 JAG09543.1 GBHO01019177 GBHO01003185 JAG24427.1 JAG40419.1 GDHC01018560 JAQ00069.1 ABLF02006304 ABLF02036268 GDHC01006567 JAQ12062.1 GBHO01023090 JAG20514.1 GBHO01027036 GBHO01010996 GBHO01010995 GBHO01010993 GBHO01010991 JAG16568.1 JAG32608.1 JAG32609.1 JAG32611.1 JAG32613.1 GBRD01007977 JAG57844.1 GBHO01006408 JAG37196.1 ABLF02017373 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 GBHO01044316 JAF99287.1 ABLF02041863 MCIT02012214 PHT96336.1 ABLF02035110 ABLF02067347 GGMR01013370 MBY25989.1 GALX01005107 JAB63359.1 GDHC01011948 JAQ06681.1 MCIT02006707 PHT96417.1 ABLF02030854 GBHO01038469 JAG05135.1 GGMR01017890 MBY30509.1 GGMR01015135 MBY27754.1 LBMM01009964 KMQ87756.1 KQ973442 KXZ75531.1 GEZM01054765 JAV73413.1 GGMR01002421 MBY15040.1 GEZM01072348 JAV65549.1 GFTR01008874 JAW07552.1 LBMM01009551 KMQ88039.1 GBBI01004409 JAC14303.1 GBBI01004407 JAC14305.1 QOIP01000009 RLU19130.1 GBHO01010487 GBHO01010485 JAG33117.1 JAG33119.1 GBHO01019080 JAG24524.1 GBHO01009080 JAG34524.1 ABLF02008636 ABLF02065931 GBHO01010489 GBHO01010484 JAG33115.1 JAG33120.1 JTDY01005490 KOB66948.1 LBMM01012392 KMQ86241.1 GBHO01034064 JAG09540.1 ABLF02008635 ABLF02023121 ABLF02065930 LNIX01000063 OXA37199.1 LBMM01018894 KMQ83668.1 GBHO01031928 GBHO01031926 JAG11676.1 JAG11678.1 ABLF02017056 LNIX01000016 OXA46198.1 LNIX01000069 OXA36894.1 GBHO01045636 GBHO01045635 JAF97967.1 JAF97968.1 LNIX01000007 OXA51795.1 GAIX01013989 JAA78571.1 RLU18850.1 GFTR01008866 JAW07560.1 ABLF02036131 ABLF02041527 ABLF02064049 ABLF02028666 ABLF02066713 GDHC01014057 JAQ04572.1 GGMS01016125 MBY85328.1 GBRD01004944 GBRD01004943 GBRD01004942 JAG60878.1 GBHO01033788 JAG09816.1 GBYB01004444 JAG74211.1
GEBQ01018232 JAT21745.1 GEBQ01015599 JAT24378.1 GECZ01006169 JAS63600.1 GECU01010337 JAS97369.1 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 GBHO01021103 GBHO01021101 JAG22501.1 JAG22503.1 ABLF02052963 GGMR01000737 MBY13356.1 GBHO01008418 GBHO01008416 JAG35186.1 JAG35188.1 GALX01004947 JAB63519.1 GBHO01004194 JAG39410.1 GEBQ01005223 JAT34754.1 GBHO01004196 JAG39408.1 ABLF02008594 GBHO01003578 JAG40026.1 GEBQ01029803 JAT10174.1 GDHC01006469 JAQ12160.1 GBHO01030122 GBHO01030121 GBHO01030120 GBHO01030116 JAG13482.1 JAG13483.1 JAG13484.1 JAG13488.1 GBHO01034065 GBHO01034061 JAG09539.1 JAG09543.1 GBHO01019177 GBHO01003185 JAG24427.1 JAG40419.1 GDHC01018560 JAQ00069.1 ABLF02006304 ABLF02036268 GDHC01006567 JAQ12062.1 GBHO01023090 JAG20514.1 GBHO01027036 GBHO01010996 GBHO01010995 GBHO01010993 GBHO01010991 JAG16568.1 JAG32608.1 JAG32609.1 JAG32611.1 JAG32613.1 GBRD01007977 JAG57844.1 GBHO01006408 JAG37196.1 ABLF02017373 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 GBHO01044316 JAF99287.1 ABLF02041863 MCIT02012214 PHT96336.1 ABLF02035110 ABLF02067347 GGMR01013370 MBY25989.1 GALX01005107 JAB63359.1 GDHC01011948 JAQ06681.1 MCIT02006707 PHT96417.1 ABLF02030854 GBHO01038469 JAG05135.1 GGMR01017890 MBY30509.1 GGMR01015135 MBY27754.1 LBMM01009964 KMQ87756.1 KQ973442 KXZ75531.1 GEZM01054765 JAV73413.1 GGMR01002421 MBY15040.1 GEZM01072348 JAV65549.1 GFTR01008874 JAW07552.1 LBMM01009551 KMQ88039.1 GBBI01004409 JAC14303.1 GBBI01004407 JAC14305.1 QOIP01000009 RLU19130.1 GBHO01010487 GBHO01010485 JAG33117.1 JAG33119.1 GBHO01019080 JAG24524.1 GBHO01009080 JAG34524.1 ABLF02008636 ABLF02065931 GBHO01010489 GBHO01010484 JAG33115.1 JAG33120.1 JTDY01005490 KOB66948.1 LBMM01012392 KMQ86241.1 GBHO01034064 JAG09540.1 ABLF02008635 ABLF02023121 ABLF02065930 LNIX01000063 OXA37199.1 LBMM01018894 KMQ83668.1 GBHO01031928 GBHO01031926 JAG11676.1 JAG11678.1 ABLF02017056 LNIX01000016 OXA46198.1 LNIX01000069 OXA36894.1 GBHO01045636 GBHO01045635 JAF97967.1 JAF97968.1 LNIX01000007 OXA51795.1 GAIX01013989 JAA78571.1 RLU18850.1 GFTR01008866 JAW07560.1 ABLF02036131 ABLF02041527 ABLF02064049 ABLF02028666 ABLF02066713 GDHC01014057 JAQ04572.1 GGMS01016125 MBY85328.1 GBRD01004944 GBRD01004943 GBRD01004942 JAG60878.1 GBHO01033788 JAG09816.1 GBYB01004444 JAG74211.1
Proteomes
Pfam
Interpro
IPR008042
Retrotrans_Pao
+ More
IPR041588 Integrase_H2C2
IPR040676 DUF5641
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR005312 DUF1759
IPR008737 Peptidase_asp_put
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR001878 Znf_CCHC
IPR041588 Integrase_H2C2
IPR040676 DUF5641
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR005312 DUF1759
IPR008737 Peptidase_asp_put
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR001878 Znf_CCHC
Gene 3D
ProteinModelPortal
Q9TYB5
O77042
D7EKN1
A0A1B6LDH0
A0A1B6LL14
A0A1B6GMH9
+ More
A0A1B6JE15 A0A1B6KZV2 A0A1B6MUZ9 A0A0A9XS77 J9M1J7 A0A2S2N9D0 A0A1S3DM66 A0A0A9YU87 V5GHM7 A0A0A9ZC74 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6MFR0 A0A0A9Z7S5 J9JVD6 A0A1S3DTZ4 A0A0A9Z7Z7 A0A1B6KG61 A0A146M0N4 A0A0A9X3X8 A0A0A9WP39 A0A0A9XUK1 A0A146KW37 X1XTX7 A0A146LZC3 A0A0A9XKM2 A0A0A9YNG0 A0A0K8SX96 A0A1S3DGE3 A0A0A9Z001 X1XMM8 A0A0A9VSF7 A0A0A9VYL4 A0A1S3DRF1 A0A1S3DNP1 J9KD60 A0A2G3AQ28 A0A3Q0JP94 X1XIA7 A0A2S2P963 V5GH76 A0A146LEU7 A0A2G3AQC0 X1WY95 A0A0A9WJX2 A0A2S2PM34 A0A1S3DM17 A0A2S2PED9 A0A0J7KBZ0 A0A1U8N896 A0A139W8E1 A0A1Y1LIG5 A0A2S2NCY5 A0A1Y1KVX6 A0A224XG39 A0A0J7KCR4 A0A023EYQ7 A0A023EYA4 A0A3L8DF94 A0A0A9YJ91 A0A0A9XX98 A0A0A9YQQ8 J9L9H5 A0A0A9YU99 A0A0L7KUL1 A0A0J7K7S0 A0A0A9WXF2 X1X412 A0A1S3DBK1 A0A1S4EIX5 A0A226CW75 A0A0J7JZS0 A0A0A9WW84 X1WSS7 A0A226DMG9 A0A226CWA5 A0A1S3DIZ2 A0A0A9VUR1 A0A226E226 S4NIN9 A0A3L8DEE4 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A224XGH3 X1WZY4 X1WYQ8 A0A146LBU6 A0A2S2R5S9 A0A0K8T5S0 A0A0A9WQV8 A0A0C9QLM0
A0A1B6JE15 A0A1B6KZV2 A0A1B6MUZ9 A0A0A9XS77 J9M1J7 A0A2S2N9D0 A0A1S3DM66 A0A0A9YU87 V5GHM7 A0A0A9ZC74 A0A3Q0IQQ3 A0A1S3CUQ4 A0A1B6MFR0 A0A0A9Z7S5 J9JVD6 A0A1S3DTZ4 A0A0A9Z7Z7 A0A1B6KG61 A0A146M0N4 A0A0A9X3X8 A0A0A9WP39 A0A0A9XUK1 A0A146KW37 X1XTX7 A0A146LZC3 A0A0A9XKM2 A0A0A9YNG0 A0A0K8SX96 A0A1S3DGE3 A0A0A9Z001 X1XMM8 A0A0A9VSF7 A0A0A9VYL4 A0A1S3DRF1 A0A1S3DNP1 J9KD60 A0A2G3AQ28 A0A3Q0JP94 X1XIA7 A0A2S2P963 V5GH76 A0A146LEU7 A0A2G3AQC0 X1WY95 A0A0A9WJX2 A0A2S2PM34 A0A1S3DM17 A0A2S2PED9 A0A0J7KBZ0 A0A1U8N896 A0A139W8E1 A0A1Y1LIG5 A0A2S2NCY5 A0A1Y1KVX6 A0A224XG39 A0A0J7KCR4 A0A023EYQ7 A0A023EYA4 A0A3L8DF94 A0A0A9YJ91 A0A0A9XX98 A0A0A9YQQ8 J9L9H5 A0A0A9YU99 A0A0L7KUL1 A0A0J7K7S0 A0A0A9WXF2 X1X412 A0A1S3DBK1 A0A1S4EIX5 A0A226CW75 A0A0J7JZS0 A0A0A9WW84 X1WSS7 A0A226DMG9 A0A226CWA5 A0A1S3DIZ2 A0A0A9VUR1 A0A226E226 S4NIN9 A0A3L8DEE4 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A224XGH3 X1WZY4 X1WYQ8 A0A146LBU6 A0A2S2R5S9 A0A0K8T5S0 A0A0A9WQV8 A0A0C9QLM0
Ontologies
Topology
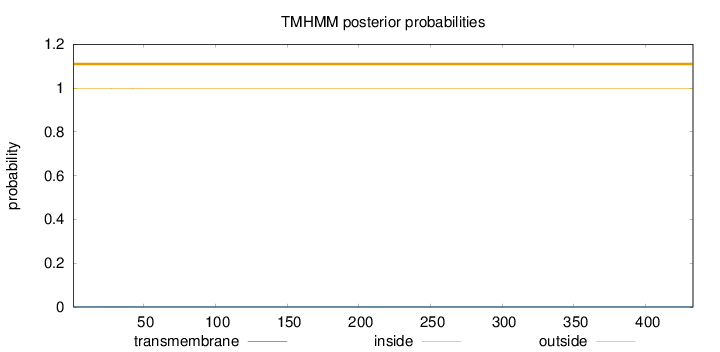
Length:
433
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01227
Exp number, first 60 AAs:
0.01143
Total prob of N-in:
0.00158
outside
1 - 433
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0.170536
CSRT
0
Interpretation
Uncertain
