Gene
KWMTBOMO08711
Pre Gene Modal
BGIBMGA007666
Annotation
PREDICTED:_phosrestin-2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.054 Nuclear Reliability : 1.333
Sequence
CDS
ATGACGTCTGACAACGCATTGAATTCGCAACGGGTGTTCAAGAAATCGTCACCCAACAATAAATTGACCTTGTACCTGGCCTCGAGGGACTTGGTGGTTGAAAACGGCAGCATCGATAGGATCCAAGGGGTGCTCCACGTGGAGGCGGATTGCTTGGAGAACAACAAGAAACTGTATGGACAAGTCACTTTGACATTCAGGTACGGTCGTGAAGATGAGGAGGTTATGGGTCTGAAGTTCTGCAACGAGGCTATCATGAGTCTCGCACAGGTCTGGCCGGTGCATTGTTCTCACGAGAGAGAGCCGAACACTCCATTACAGGATGCATTAATAAAACGCCTCGGAGCCAACGCGTTTCCTTTCCACTTGGAGCTGACTCCACTGGCGCCGCCAAGCGTCCAACTTGTACCCGCCAAGCAGTACCACGGCGCCCCCATTGGAACCTCATACGACGTTCGAGCATATGTTGCAAGGGCGGGTCGCTAG
Protein
MTSDNALNSQRVFKKSSPNNKLTLYLASRDLVVENGSIDRIQGVLHVEADCLENNKKLYGQVTLTFRYGREDEEVMGLKFCNEAIMSLAQVWPVHCSHEREPNTPLQDALIKRLGANAFPFHLELTPLAPPSVQLVPAKQYHGAPIGTSYDVRAYVARAGR
Summary
Similarity
Belongs to the heat shock protein 70 family.
Uniprot
H9JDR9
A0A2W1BJ31
A0A194PR53
A0A212F365
A0A437BSA5
A0A194QM58
+ More
D6X567 A0A1Y1KDL0 A0A067QZW4 A0A088ACD6 A0A026W496 A0A3L8E1H1 A0A154PI05 A0A2S2NP95 J9JQG1 A0A2H8TLP6 K7J8Y4 A0A0L7R5F4 A0A182M5K4 E0VU89 A0A182T175 A0A182YHG9 A0A182VWL2 A0A182X0A1 F4W612 A0A182P576 A0A182VIA8 A0A182LR30 A0A182HZZ1 A0A1A9YM57 A0A158P2I6 A0A182TT08 A0A195EQ91 A0A182FNZ2 A0A1J1IQN1 A0A1B0A1V5 A0A182QLY4 A0A151J9U0 A0A1S4GEG1 W5JJT1 A0A151ICZ6 A0A182R4S9 Q7PUI7 A0A1B0G9U6 A0A1B0C185 B1Q294 A0A195AWE8 A0A1I8PVM8 A0A1A9UGJ2 A0A1A9W1Y5 A0A0L0CH99 A0A336MCB4 A0A1I8MJP5 A0A336MDF7 W8B871 B4JKQ6 B4L2E8 B4MT53 B4IDQ5 Q6NN64 B4MEF1 A0A1W4V7J3 A0A0Q5T3T1 B3NW30 B4PXI9 Q9W2V2 B3MY16 B4H0C3 A0A0R3P091 Q29H45 A0A0C9RKZ9 A0A1B6MP72 A0A1B0CBA9 A0A182N5U5 A0A0M9A795 A0A232FB99 K7IUT7 A0A0L7RIZ4 B7PWE0 A0A0P5PCE9 A0A0P5D6I8 A0A1B6LYY3 A0A0P5TQD4 A0A0P5Z5N6 A0A0N8E0J8 A0A0N8C5E2 A0A0P5GKR8 A0A0P4XLF1 A0A0P4XHY9 A0A0P5LQZ4 D1MVD8 A0A0N8A9L3 A0A0P4ZIE8
D6X567 A0A1Y1KDL0 A0A067QZW4 A0A088ACD6 A0A026W496 A0A3L8E1H1 A0A154PI05 A0A2S2NP95 J9JQG1 A0A2H8TLP6 K7J8Y4 A0A0L7R5F4 A0A182M5K4 E0VU89 A0A182T175 A0A182YHG9 A0A182VWL2 A0A182X0A1 F4W612 A0A182P576 A0A182VIA8 A0A182LR30 A0A182HZZ1 A0A1A9YM57 A0A158P2I6 A0A182TT08 A0A195EQ91 A0A182FNZ2 A0A1J1IQN1 A0A1B0A1V5 A0A182QLY4 A0A151J9U0 A0A1S4GEG1 W5JJT1 A0A151ICZ6 A0A182R4S9 Q7PUI7 A0A1B0G9U6 A0A1B0C185 B1Q294 A0A195AWE8 A0A1I8PVM8 A0A1A9UGJ2 A0A1A9W1Y5 A0A0L0CH99 A0A336MCB4 A0A1I8MJP5 A0A336MDF7 W8B871 B4JKQ6 B4L2E8 B4MT53 B4IDQ5 Q6NN64 B4MEF1 A0A1W4V7J3 A0A0Q5T3T1 B3NW30 B4PXI9 Q9W2V2 B3MY16 B4H0C3 A0A0R3P091 Q29H45 A0A0C9RKZ9 A0A1B6MP72 A0A1B0CBA9 A0A182N5U5 A0A0M9A795 A0A232FB99 K7IUT7 A0A0L7RIZ4 B7PWE0 A0A0P5PCE9 A0A0P5D6I8 A0A1B6LYY3 A0A0P5TQD4 A0A0P5Z5N6 A0A0N8E0J8 A0A0N8C5E2 A0A0P5GKR8 A0A0P4XLF1 A0A0P4XHY9 A0A0P5LQZ4 D1MVD8 A0A0N8A9L3 A0A0P4ZIE8
Pubmed
19121390
28756777
26354079
22118469
18362917
19820115
+ More
28004739 24845553 24508170 30249741 20075255 20566863 25244985 21719571 20966253 21347285 12364791 20920257 23761445 14986925 26108605 25315136 24495485 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23185243 28648823 20347048
28004739 24845553 24508170 30249741 20075255 20566863 25244985 21719571 20966253 21347285 12364791 20920257 23761445 14986925 26108605 25315136 24495485 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23185243 28648823 20347048
EMBL
BABH01003088
BABH01003089
KZ150012
PZC75049.1
KQ459596
KPI95438.1
+ More
AGBW02010613 OWR48177.1 RSAL01000014 RVE53225.1 KQ461195 KPJ06628.1 KQ971382 EEZ97692.2 GEZM01086398 GEZM01086397 JAV59484.1 KK853055 KDR11977.1 KK107436 EZA50838.1 QOIP01000002 RLU26039.1 KQ434923 KZC11485.1 GGMR01006391 MBY19010.1 ABLF02027463 ABLF02027464 GFXV01002787 MBW14592.1 KQ414654 KOC66054.1 AXCM01005645 AXCM01005646 DS235783 EEB16945.1 GL887707 EGI70147.1 APCN01002026 ADTU01006876 ADTU01006877 ADTU01006878 ADTU01006879 ADTU01006880 ADTU01006881 ADTU01006882 ADTU01006883 KQ982021 KYN30450.1 CVRI01000057 CRL02050.1 AXCN02001153 KQ979361 KYN21804.1 AAAB01008987 ADMH02001287 ETN63160.1 KQ977995 KYM98262.1 EAA01439.5 CCAG010011662 JXJN01023903 BK001417 DAA01945.1 KQ976731 KYM76360.1 JRES01000406 KNC31625.1 UFQS01000685 UFQT01000685 SSX06205.1 SSX26559.1 UFQT01000991 SSX28325.1 GAMC01011723 JAB94832.1 CH916370 EDW00159.1 CH933810 EDW06824.1 KRF93827.1 KRF93828.1 CH963851 EDW75292.2 CH480830 EDW45713.1 BT011430 AAR99088.1 CH940664 EDW62926.2 CH954180 KQS29855.1 EDV46163.1 CM000162 EDX02940.1 KRK07029.1 KRK07030.1 KRK07031.1 KRK07032.1 KRK07033.1 AE014298 BT133472 AAF46586.2 AFH97162.1 CH902630 EDV38631.1 CH479200 EDW29718.1 CH379064 KRT06673.1 EAL31913.3 KRT06671.1 GBYB01004777 GBYB01013246 GBYB01013997 JAG74544.1 JAG83013.1 JAG83764.1 GEBQ01002221 JAT37756.1 AJWK01004942 KQ435727 KOX77906.1 NNAY01000550 OXU27729.1 AAZX01002635 KQ414583 KOC70716.1 ABJB010030301 ABJB010502265 ABJB010770397 ABJB011055887 DS807600 EEC10912.1 GDIQ01130637 JAL21089.1 GDIP01176334 JAJ47068.1 GEBQ01011163 JAT28814.1 GDIP01123425 JAL80289.1 GDIP01061293 JAM42422.1 GDIQ01073684 JAN21053.1 GDIQ01113123 JAL38603.1 GDIQ01242985 JAK08740.1 GDIP01240930 JAI82471.1 GDIP01240931 JAI82470.1 GDIQ01168517 JAK83208.1 AB293440 BAI50912.1 GDIP01169033 JAJ54369.1 GDIP01225578 JAI97823.1
AGBW02010613 OWR48177.1 RSAL01000014 RVE53225.1 KQ461195 KPJ06628.1 KQ971382 EEZ97692.2 GEZM01086398 GEZM01086397 JAV59484.1 KK853055 KDR11977.1 KK107436 EZA50838.1 QOIP01000002 RLU26039.1 KQ434923 KZC11485.1 GGMR01006391 MBY19010.1 ABLF02027463 ABLF02027464 GFXV01002787 MBW14592.1 KQ414654 KOC66054.1 AXCM01005645 AXCM01005646 DS235783 EEB16945.1 GL887707 EGI70147.1 APCN01002026 ADTU01006876 ADTU01006877 ADTU01006878 ADTU01006879 ADTU01006880 ADTU01006881 ADTU01006882 ADTU01006883 KQ982021 KYN30450.1 CVRI01000057 CRL02050.1 AXCN02001153 KQ979361 KYN21804.1 AAAB01008987 ADMH02001287 ETN63160.1 KQ977995 KYM98262.1 EAA01439.5 CCAG010011662 JXJN01023903 BK001417 DAA01945.1 KQ976731 KYM76360.1 JRES01000406 KNC31625.1 UFQS01000685 UFQT01000685 SSX06205.1 SSX26559.1 UFQT01000991 SSX28325.1 GAMC01011723 JAB94832.1 CH916370 EDW00159.1 CH933810 EDW06824.1 KRF93827.1 KRF93828.1 CH963851 EDW75292.2 CH480830 EDW45713.1 BT011430 AAR99088.1 CH940664 EDW62926.2 CH954180 KQS29855.1 EDV46163.1 CM000162 EDX02940.1 KRK07029.1 KRK07030.1 KRK07031.1 KRK07032.1 KRK07033.1 AE014298 BT133472 AAF46586.2 AFH97162.1 CH902630 EDV38631.1 CH479200 EDW29718.1 CH379064 KRT06673.1 EAL31913.3 KRT06671.1 GBYB01004777 GBYB01013246 GBYB01013997 JAG74544.1 JAG83013.1 JAG83764.1 GEBQ01002221 JAT37756.1 AJWK01004942 KQ435727 KOX77906.1 NNAY01000550 OXU27729.1 AAZX01002635 KQ414583 KOC70716.1 ABJB010030301 ABJB010502265 ABJB010770397 ABJB011055887 DS807600 EEC10912.1 GDIQ01130637 JAL21089.1 GDIP01176334 JAJ47068.1 GEBQ01011163 JAT28814.1 GDIP01123425 JAL80289.1 GDIP01061293 JAM42422.1 GDIQ01073684 JAN21053.1 GDIQ01113123 JAL38603.1 GDIQ01242985 JAK08740.1 GDIP01240930 JAI82471.1 GDIP01240931 JAI82470.1 GDIQ01168517 JAK83208.1 AB293440 BAI50912.1 GDIP01169033 JAJ54369.1 GDIP01225578 JAI97823.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000283053
UP000053240
UP000007266
+ More
UP000027135 UP000005203 UP000053097 UP000279307 UP000076502 UP000007819 UP000002358 UP000053825 UP000075883 UP000009046 UP000075901 UP000076408 UP000075920 UP000076407 UP000007755 UP000075885 UP000075903 UP000075882 UP000075840 UP000092443 UP000005205 UP000075902 UP000078541 UP000069272 UP000183832 UP000092445 UP000075886 UP000078492 UP000000673 UP000078542 UP000075900 UP000007062 UP000092444 UP000092460 UP000078540 UP000095300 UP000078200 UP000091820 UP000037069 UP000095301 UP000001070 UP000009192 UP000007798 UP000001292 UP000008792 UP000192221 UP000008711 UP000002282 UP000000803 UP000007801 UP000008744 UP000001819 UP000092461 UP000075884 UP000053105 UP000215335 UP000001555
UP000027135 UP000005203 UP000053097 UP000279307 UP000076502 UP000007819 UP000002358 UP000053825 UP000075883 UP000009046 UP000075901 UP000076408 UP000075920 UP000076407 UP000007755 UP000075885 UP000075903 UP000075882 UP000075840 UP000092443 UP000005205 UP000075902 UP000078541 UP000069272 UP000183832 UP000092445 UP000075886 UP000078492 UP000000673 UP000078542 UP000075900 UP000007062 UP000092444 UP000092460 UP000078540 UP000095300 UP000078200 UP000091820 UP000037069 UP000095301 UP000001070 UP000009192 UP000007798 UP000001292 UP000008792 UP000192221 UP000008711 UP000002282 UP000000803 UP000007801 UP000008744 UP000001819 UP000092461 UP000075884 UP000053105 UP000215335 UP000001555
Interpro
ProteinModelPortal
H9JDR9
A0A2W1BJ31
A0A194PR53
A0A212F365
A0A437BSA5
A0A194QM58
+ More
D6X567 A0A1Y1KDL0 A0A067QZW4 A0A088ACD6 A0A026W496 A0A3L8E1H1 A0A154PI05 A0A2S2NP95 J9JQG1 A0A2H8TLP6 K7J8Y4 A0A0L7R5F4 A0A182M5K4 E0VU89 A0A182T175 A0A182YHG9 A0A182VWL2 A0A182X0A1 F4W612 A0A182P576 A0A182VIA8 A0A182LR30 A0A182HZZ1 A0A1A9YM57 A0A158P2I6 A0A182TT08 A0A195EQ91 A0A182FNZ2 A0A1J1IQN1 A0A1B0A1V5 A0A182QLY4 A0A151J9U0 A0A1S4GEG1 W5JJT1 A0A151ICZ6 A0A182R4S9 Q7PUI7 A0A1B0G9U6 A0A1B0C185 B1Q294 A0A195AWE8 A0A1I8PVM8 A0A1A9UGJ2 A0A1A9W1Y5 A0A0L0CH99 A0A336MCB4 A0A1I8MJP5 A0A336MDF7 W8B871 B4JKQ6 B4L2E8 B4MT53 B4IDQ5 Q6NN64 B4MEF1 A0A1W4V7J3 A0A0Q5T3T1 B3NW30 B4PXI9 Q9W2V2 B3MY16 B4H0C3 A0A0R3P091 Q29H45 A0A0C9RKZ9 A0A1B6MP72 A0A1B0CBA9 A0A182N5U5 A0A0M9A795 A0A232FB99 K7IUT7 A0A0L7RIZ4 B7PWE0 A0A0P5PCE9 A0A0P5D6I8 A0A1B6LYY3 A0A0P5TQD4 A0A0P5Z5N6 A0A0N8E0J8 A0A0N8C5E2 A0A0P5GKR8 A0A0P4XLF1 A0A0P4XHY9 A0A0P5LQZ4 D1MVD8 A0A0N8A9L3 A0A0P4ZIE8
D6X567 A0A1Y1KDL0 A0A067QZW4 A0A088ACD6 A0A026W496 A0A3L8E1H1 A0A154PI05 A0A2S2NP95 J9JQG1 A0A2H8TLP6 K7J8Y4 A0A0L7R5F4 A0A182M5K4 E0VU89 A0A182T175 A0A182YHG9 A0A182VWL2 A0A182X0A1 F4W612 A0A182P576 A0A182VIA8 A0A182LR30 A0A182HZZ1 A0A1A9YM57 A0A158P2I6 A0A182TT08 A0A195EQ91 A0A182FNZ2 A0A1J1IQN1 A0A1B0A1V5 A0A182QLY4 A0A151J9U0 A0A1S4GEG1 W5JJT1 A0A151ICZ6 A0A182R4S9 Q7PUI7 A0A1B0G9U6 A0A1B0C185 B1Q294 A0A195AWE8 A0A1I8PVM8 A0A1A9UGJ2 A0A1A9W1Y5 A0A0L0CH99 A0A336MCB4 A0A1I8MJP5 A0A336MDF7 W8B871 B4JKQ6 B4L2E8 B4MT53 B4IDQ5 Q6NN64 B4MEF1 A0A1W4V7J3 A0A0Q5T3T1 B3NW30 B4PXI9 Q9W2V2 B3MY16 B4H0C3 A0A0R3P091 Q29H45 A0A0C9RKZ9 A0A1B6MP72 A0A1B0CBA9 A0A182N5U5 A0A0M9A795 A0A232FB99 K7IUT7 A0A0L7RIZ4 B7PWE0 A0A0P5PCE9 A0A0P5D6I8 A0A1B6LYY3 A0A0P5TQD4 A0A0P5Z5N6 A0A0N8E0J8 A0A0N8C5E2 A0A0P5GKR8 A0A0P4XLF1 A0A0P4XHY9 A0A0P5LQZ4 D1MVD8 A0A0N8A9L3 A0A0P4ZIE8
PDB
6BK9
E-value=1.02349e-25,
Score=284
Ontologies
GO
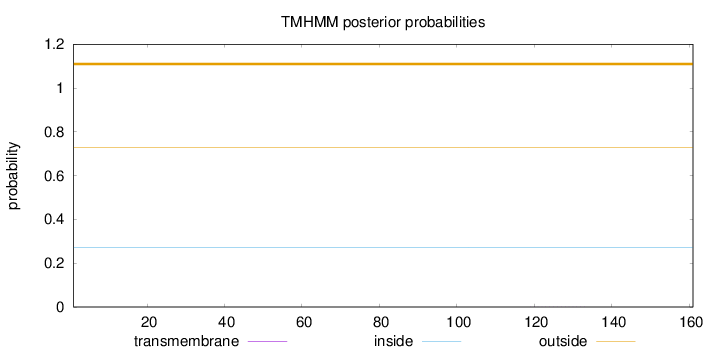
Topology
Length:
161
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00403
Exp number, first 60 AAs:
0
Total prob of N-in:
0.27054
outside
1 - 161
Population Genetic Test Statistics
Pi
192.233725
Theta
148.823995
Tajima's D
0.248652
CLR
0.094229
CSRT
0.44242787860607
Interpretation
Uncertain
