Gene
KWMTBOMO08709 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007920
Annotation
PREDICTED:_chondroitin_proteoglycan-2-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 4.278
Sequence
CDS
ATGTGGTTCAGCCCTACCCTTCAGCGTGTCTATAGAGAACATTCCCCTCTTTGTTTGCTGGAGCATAAATCCTTAAGCGTGATCCTGGCAGCCATAAACCTAACATCATGTGCCCAAATCGAGGACCCGTGCAAGACCAAAGCTAGAATAGTGGCCGATGACAAATACTGCGACAAGTATTGGGAATGTGACAACGGACAATCGGTACAATACGACTGCCCCAATGGACTTGTTTTTGCGGGCAAACATCGAGGGGTGACTGAGGGCTGCGATTACCCTTGGAGGTCAAATTACTGCGAGTACCCGAAGGCTCAGATAAATCCCCCAATCGGCACGGAGCACTGCGACTGGCTGTACGGTATCTTTGGCCACGAGACCTCCTGCACCAGGTACTGGACCTGCTGGAACGGCACCGCCACGGAGCAGCTTTGCATCGGAGGGCTTTTGTACAATGAGAACGCCCACTCTTGTGACTGGCCTGAGAACGTTGACGGTTGTCAAAAGCATCCTCTCTGCAACGAGGACCCGAACGGTAACGTGCCCTTAGGCAAGTCCTGCAACCGGTACTGGCAGTGCCAGGGCGGATATCCTCGGCTGCAGCGTTGCCCTGCTATGTTGGTGTTTGACAGGCGCTCGCTGAGATGCGTGGTACCACCCACCGAGGACTGCGACATCCCCACCACCACCGCTCCACCACCAGAAGAAGAGCAGCAGGATGCGCGACAGGGACAACACCAGAAGCGGCCCTCCAACGATTTCCAAGAGCAGCAACAGGGCCGCACAAGACCGAGGAACTAA
Protein
MWFSPTLQRVYREHSPLCLLEHKSLSVILAAINLTSCAQIEDPCKTKARIVADDKYCDKYWECDNGQSVQYDCPNGLVFAGKHRGVTEGCDYPWRSNYCEYPKAQINPPIGTEHCDWLYGIFGHETSCTRYWTCWNGTATEQLCIGGLLYNENAHSCDWPENVDGCQKHPLCNEDPNGNVPLGKSCNRYWQCQGGYPRLQRCPAMLVFDRRSLRCVVPPTEDCDIPTTTAPPPEEEQQDARQGQHQKRPSNDFQEQQQGRTRPRN
Summary
Uniprot
A0A2A4JUL9
A0A2W1BJP1
A0A194PQ45
I4DJ00
I4DM84
A0A2H1WCP6
+ More
H9JEH3 J3JV56 A0A411G6L7 A0A182XHH9 A0A0C9RJ66 A1YR30 A0A0L0CML4 A0A1B6CQM1 T1PAP8 A0A1I8MUE2 A0A3L8E5I7 K7IMV0 A0A2S1ZS44 A0A1B0A3C3 W8AWR9 A0A0K8V7P5 A0A195FLS9 A0A026WEA2 A0A1A9VBU7 A0A158NER6 A0A034W9H0 E2A162 A0A1Y1KUZ8 A0A0A1WZW3 A0A2J7QFM3 A0A1B0FRA3 A0A0M9A9K6 E0VTP1 A0A1D1Y2N2 A0A0L7QP97 A0A3S7SJV9 A0A067R5M3 A0A154PBC4 A0A1B0DG13 A0A1Y1KS78 E2B780 A0A087ZR63 A0A310SFW2 A0A1Y1KVS0 A0A023F9L5 A0A1A9WLR4 A0A1B6MNB4 Q5TU29 A0A182HTW7 A0A2S2PCP1 A0A069DRR5 R4FP21 A0A0J7KRJ3 A0A195DZQ9 A0A151XIS6 A0A224XUL7 W5JF71 E9IAA4 J9JMX9 A0A195AVQ7 A0A195CRB6 A0A2S2QB74 A0A023H838 A0A0P4W2Y4 A0A164WRK2 A0A0P5RHW0 A0A0P5WKF6 A0A0P5DDP2 A0A0N8DEB1 A0A0P5HB82 A0A0N8C9B9 A0A0P5GNU4 A0A0P5JSN8 A0A0N8D9Y4 A0A0P5DXW3 A0A0P5IKJ7 A0A0P5UCX7
H9JEH3 J3JV56 A0A411G6L7 A0A182XHH9 A0A0C9RJ66 A1YR30 A0A0L0CML4 A0A1B6CQM1 T1PAP8 A0A1I8MUE2 A0A3L8E5I7 K7IMV0 A0A2S1ZS44 A0A1B0A3C3 W8AWR9 A0A0K8V7P5 A0A195FLS9 A0A026WEA2 A0A1A9VBU7 A0A158NER6 A0A034W9H0 E2A162 A0A1Y1KUZ8 A0A0A1WZW3 A0A2J7QFM3 A0A1B0FRA3 A0A0M9A9K6 E0VTP1 A0A1D1Y2N2 A0A0L7QP97 A0A3S7SJV9 A0A067R5M3 A0A154PBC4 A0A1B0DG13 A0A1Y1KS78 E2B780 A0A087ZR63 A0A310SFW2 A0A1Y1KVS0 A0A023F9L5 A0A1A9WLR4 A0A1B6MNB4 Q5TU29 A0A182HTW7 A0A2S2PCP1 A0A069DRR5 R4FP21 A0A0J7KRJ3 A0A195DZQ9 A0A151XIS6 A0A224XUL7 W5JF71 E9IAA4 J9JMX9 A0A195AVQ7 A0A195CRB6 A0A2S2QB74 A0A023H838 A0A0P4W2Y4 A0A164WRK2 A0A0P5RHW0 A0A0P5WKF6 A0A0P5DDP2 A0A0N8DEB1 A0A0P5HB82 A0A0N8C9B9 A0A0P5GNU4 A0A0P5JSN8 A0A0N8D9Y4 A0A0P5DXW3 A0A0P5IKJ7 A0A0P5UCX7
Pubmed
EMBL
NWSH01000550
PCG75701.1
KZ150142
PZC72940.1
KQ459596
KPI95437.1
+ More
AK401268 BAM17890.1 AK402402 BAM19024.1 ODYU01007783 SOQ50840.1 BABH01003098 APGK01017227 BT127122 KB739998 KB631984 AEE62084.1 ENN81971.1 ERL87661.1 MH365717 QBB01526.1 GBYB01002879 GBYB01013267 JAG72646.1 JAG83034.1 EF125547 KQ971316 ABL73931.1 EEZ98786.1 JRES01000292 KNC32684.1 GEDC01021558 JAS15740.1 KA644988 AFP59617.1 QOIP01000001 RLU27138.1 MF942765 AWK28288.1 GAMC01016018 GAMC01016017 JAB90537.1 GDHF01027495 GDHF01017362 JAI24819.1 JAI34952.1 KQ981490 KYN41286.1 KK107260 EZA53996.1 ADTU01013616 ADTU01013617 ADTU01013618 GAKP01007985 JAC50967.1 GL435707 EFN72829.1 GEZM01075112 JAV64271.1 GBXI01010869 GBXI01010222 GBXI01001404 GBXI01001248 JAD03423.1 JAD04070.1 JAD12888.1 JAD13044.1 NEVH01015297 PNF27343.1 CCAG010016939 CCAG010016940 KQ435709 KOX80017.1 DS235768 EEB16747.1 GDJX01019055 JAT48881.1 KQ414821 KOC60452.1 MG601722 AYA50047.1 KK852869 KDR14642.1 KQ434867 KZC09152.1 AJVK01033546 GEZM01075115 JAV64269.1 GL446149 EFN88441.1 KQ773121 OAD52430.1 GEZM01075113 JAV64270.1 GBBI01001099 JAC17613.1 GEBQ01002547 JAT37430.1 AAAB01008859 EAL40816.2 APCN01000586 GGMR01014591 MBY27210.1 GBGD01002259 JAC86630.1 GAHY01000338 JAA77172.1 LBMM01003945 KMQ92963.1 KQ979999 KYN18316.1 KQ982080 KYQ60237.1 GFTR01004757 JAW11669.1 ADMH02001371 ETN62736.1 GL761997 EFZ22492.1 ABLF02036175 KQ976736 KYM76110.1 KQ977381 KYN03180.1 GGMS01005577 MBY74780.1 KC200273 AGG20312.1 GDRN01076702 JAI62868.1 LRGB01001126 KZS13507.1 GDIQ01100557 JAL51169.1 GDIP01097865 JAM05850.1 GDIP01161467 JAJ61935.1 GDIP01042159 JAM61556.1 GDIQ01235498 JAK16227.1 GDIQ01102107 JAL49619.1 GDIQ01237760 JAK13965.1 GDIQ01220066 JAK31659.1 GDIP01054375 JAM49340.1 GDIP01150909 JAJ72493.1 GDIQ01220065 JAK31660.1 GDIP01114864 JAL88850.1
AK401268 BAM17890.1 AK402402 BAM19024.1 ODYU01007783 SOQ50840.1 BABH01003098 APGK01017227 BT127122 KB739998 KB631984 AEE62084.1 ENN81971.1 ERL87661.1 MH365717 QBB01526.1 GBYB01002879 GBYB01013267 JAG72646.1 JAG83034.1 EF125547 KQ971316 ABL73931.1 EEZ98786.1 JRES01000292 KNC32684.1 GEDC01021558 JAS15740.1 KA644988 AFP59617.1 QOIP01000001 RLU27138.1 MF942765 AWK28288.1 GAMC01016018 GAMC01016017 JAB90537.1 GDHF01027495 GDHF01017362 JAI24819.1 JAI34952.1 KQ981490 KYN41286.1 KK107260 EZA53996.1 ADTU01013616 ADTU01013617 ADTU01013618 GAKP01007985 JAC50967.1 GL435707 EFN72829.1 GEZM01075112 JAV64271.1 GBXI01010869 GBXI01010222 GBXI01001404 GBXI01001248 JAD03423.1 JAD04070.1 JAD12888.1 JAD13044.1 NEVH01015297 PNF27343.1 CCAG010016939 CCAG010016940 KQ435709 KOX80017.1 DS235768 EEB16747.1 GDJX01019055 JAT48881.1 KQ414821 KOC60452.1 MG601722 AYA50047.1 KK852869 KDR14642.1 KQ434867 KZC09152.1 AJVK01033546 GEZM01075115 JAV64269.1 GL446149 EFN88441.1 KQ773121 OAD52430.1 GEZM01075113 JAV64270.1 GBBI01001099 JAC17613.1 GEBQ01002547 JAT37430.1 AAAB01008859 EAL40816.2 APCN01000586 GGMR01014591 MBY27210.1 GBGD01002259 JAC86630.1 GAHY01000338 JAA77172.1 LBMM01003945 KMQ92963.1 KQ979999 KYN18316.1 KQ982080 KYQ60237.1 GFTR01004757 JAW11669.1 ADMH02001371 ETN62736.1 GL761997 EFZ22492.1 ABLF02036175 KQ976736 KYM76110.1 KQ977381 KYN03180.1 GGMS01005577 MBY74780.1 KC200273 AGG20312.1 GDRN01076702 JAI62868.1 LRGB01001126 KZS13507.1 GDIQ01100557 JAL51169.1 GDIP01097865 JAM05850.1 GDIP01161467 JAJ61935.1 GDIP01042159 JAM61556.1 GDIQ01235498 JAK16227.1 GDIQ01102107 JAL49619.1 GDIQ01237760 JAK13965.1 GDIQ01220066 JAK31659.1 GDIP01054375 JAM49340.1 GDIP01150909 JAJ72493.1 GDIQ01220065 JAK31660.1 GDIP01114864 JAL88850.1
Proteomes
UP000218220
UP000053268
UP000005204
UP000019118
UP000030742
UP000076407
+ More
UP000007266 UP000037069 UP000095301 UP000279307 UP000002358 UP000092445 UP000078541 UP000053097 UP000078200 UP000005205 UP000000311 UP000235965 UP000092444 UP000053105 UP000009046 UP000053825 UP000027135 UP000076502 UP000092462 UP000008237 UP000005203 UP000091820 UP000007062 UP000075840 UP000036403 UP000078492 UP000075809 UP000000673 UP000007819 UP000078540 UP000078542 UP000076858
UP000007266 UP000037069 UP000095301 UP000279307 UP000002358 UP000092445 UP000078541 UP000053097 UP000078200 UP000005205 UP000000311 UP000235965 UP000092444 UP000053105 UP000009046 UP000053825 UP000027135 UP000076502 UP000092462 UP000008237 UP000005203 UP000091820 UP000007062 UP000075840 UP000036403 UP000078492 UP000075809 UP000000673 UP000007819 UP000078540 UP000078542 UP000076858
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JUL9
A0A2W1BJP1
A0A194PQ45
I4DJ00
I4DM84
A0A2H1WCP6
+ More
H9JEH3 J3JV56 A0A411G6L7 A0A182XHH9 A0A0C9RJ66 A1YR30 A0A0L0CML4 A0A1B6CQM1 T1PAP8 A0A1I8MUE2 A0A3L8E5I7 K7IMV0 A0A2S1ZS44 A0A1B0A3C3 W8AWR9 A0A0K8V7P5 A0A195FLS9 A0A026WEA2 A0A1A9VBU7 A0A158NER6 A0A034W9H0 E2A162 A0A1Y1KUZ8 A0A0A1WZW3 A0A2J7QFM3 A0A1B0FRA3 A0A0M9A9K6 E0VTP1 A0A1D1Y2N2 A0A0L7QP97 A0A3S7SJV9 A0A067R5M3 A0A154PBC4 A0A1B0DG13 A0A1Y1KS78 E2B780 A0A087ZR63 A0A310SFW2 A0A1Y1KVS0 A0A023F9L5 A0A1A9WLR4 A0A1B6MNB4 Q5TU29 A0A182HTW7 A0A2S2PCP1 A0A069DRR5 R4FP21 A0A0J7KRJ3 A0A195DZQ9 A0A151XIS6 A0A224XUL7 W5JF71 E9IAA4 J9JMX9 A0A195AVQ7 A0A195CRB6 A0A2S2QB74 A0A023H838 A0A0P4W2Y4 A0A164WRK2 A0A0P5RHW0 A0A0P5WKF6 A0A0P5DDP2 A0A0N8DEB1 A0A0P5HB82 A0A0N8C9B9 A0A0P5GNU4 A0A0P5JSN8 A0A0N8D9Y4 A0A0P5DXW3 A0A0P5IKJ7 A0A0P5UCX7
H9JEH3 J3JV56 A0A411G6L7 A0A182XHH9 A0A0C9RJ66 A1YR30 A0A0L0CML4 A0A1B6CQM1 T1PAP8 A0A1I8MUE2 A0A3L8E5I7 K7IMV0 A0A2S1ZS44 A0A1B0A3C3 W8AWR9 A0A0K8V7P5 A0A195FLS9 A0A026WEA2 A0A1A9VBU7 A0A158NER6 A0A034W9H0 E2A162 A0A1Y1KUZ8 A0A0A1WZW3 A0A2J7QFM3 A0A1B0FRA3 A0A0M9A9K6 E0VTP1 A0A1D1Y2N2 A0A0L7QP97 A0A3S7SJV9 A0A067R5M3 A0A154PBC4 A0A1B0DG13 A0A1Y1KS78 E2B780 A0A087ZR63 A0A310SFW2 A0A1Y1KVS0 A0A023F9L5 A0A1A9WLR4 A0A1B6MNB4 Q5TU29 A0A182HTW7 A0A2S2PCP1 A0A069DRR5 R4FP21 A0A0J7KRJ3 A0A195DZQ9 A0A151XIS6 A0A224XUL7 W5JF71 E9IAA4 J9JMX9 A0A195AVQ7 A0A195CRB6 A0A2S2QB74 A0A023H838 A0A0P4W2Y4 A0A164WRK2 A0A0P5RHW0 A0A0P5WKF6 A0A0P5DDP2 A0A0N8DEB1 A0A0P5HB82 A0A0N8C9B9 A0A0P5GNU4 A0A0P5JSN8 A0A0N8D9Y4 A0A0P5DXW3 A0A0P5IKJ7 A0A0P5UCX7
PDB
5WVG
E-value=4.85686e-06,
Score=118
Ontologies
GO
PANTHER
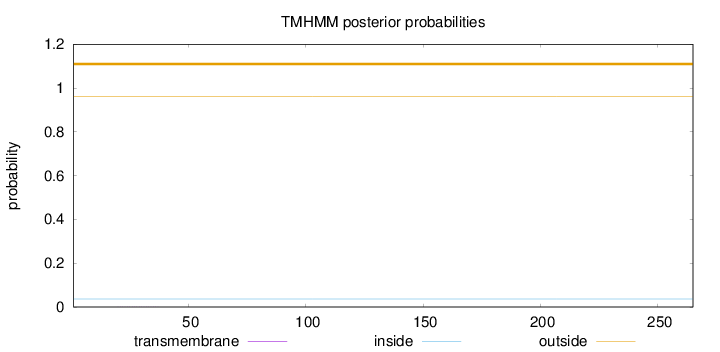
Topology
Length:
265
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00410999999999999
Exp number, first 60 AAs:
0.00391
Total prob of N-in:
0.03696
outside
1 - 265
Population Genetic Test Statistics
Pi
191.738159
Theta
172.267702
Tajima's D
0.316445
CLR
0.544008
CSRT
0.459877006149693
Interpretation
Uncertain
