Gene
KWMTBOMO08708 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007921
Annotation
PREDICTED:_kynurenine--oxoglutarate_transaminase_3_isoform_X1_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 4.095
Sequence
CDS
ATGGCAGAAAAGTTTCGATTGCCGGAACGTTATGGAGCTGGTGAGAAAAGCGTATGGGTTGAGTACATACAATTAGCTGCAGAGTATAAGCCTGCCGTGAACCTTGGTCAGGGCTTTCCTGACTATCACGCACCGAAACATGTAACAGAAGCACTATCGCAGATAGCTACCAGTGAAAATCCACTGCTTCATCAGTACACAAGAGGCTTTGGTCTGCCGCGCCTCGTGGAGAATCTATCAAAGGTTTACTCTCCGCTGATTGGACGACAGATAGATGCCTTCAATGAAATCCTAGTCACGAGCGGTGCCTATGAAGCACTTTACTCTACCATTTTAGGTCACGTGGATACCGGGGATGAGGTGATCATTATAGAACCATATTTCGACTGCTACGACTTCATGGTGAAGTGCGCCGGAGGTGTACCTAGGTTCATCGCTCTGAAACCGAAACCTCAAGGCGACGATATATCATCAGCGGATTGGGTTTTAGACGAGGCGGAGCTAGCTTCGCTCTTCAACAACCGAACTAAAATGATCATCGTGAACACGCCCCACAATCCGCTGGGCAAGGTGTTCACCCAACGCGAGCTGGAACTCATTGCCGATCTGTGCAAGAAGCACAATGTGCTTTGTCTCTCCGACGAAGTCTACGAGTGGATGGTGTACGAGCCCAAACAGCATATCCGGATAGCGTCATTCCCGGGCATGTGGGAGAGGACCATCACCGTCGGGTCCGCGGGGAAGACGTTCTCCGTCACCGGGTGGAAGATCGGGTGGGCGTACGGACCAGCCGACCTGATGAGGAATCTGCAAGTGGTACACCAGAACTGTGTGTACACGTGCTGCACGCCGGTCGAGGAAGCCGTGGCCCGCTCGTTCGAGTACGAGCTGGCCAGGCGCGACTCGCCGGACTGCTACTTCTATTCTCTGGCTCGAGAGCTGCGCCCGAAGCGAGACTACCTCATGAAGATCTTGAAGGAGAACGGCTTCAAACCGACCCTACCCGAGGCCGGGTACTTTGTCGTGGCCGATTGGACTGACTTGGAAAAGAAGATCGACTTATCGAGCGAACTGGACAAATACAAAGATTACAAGTTCACAAAGAAATTCGCTAAAGAGGCGGGCGTACTCGCCATACCGCCGAGCGCCTTCTACTCTGAAGCACACAAACACCTGGGGGAGACGTTCGCGAGATTCTGCTTTATAAAGAAAGACGAAAATCTAGAACTAACCGAGAATCTGCTAAAGGAATGGAATTCTAAGAAAAAGTAA
Protein
MAEKFRLPERYGAGEKSVWVEYIQLAAEYKPAVNLGQGFPDYHAPKHVTEALSQIATSENPLLHQYTRGFGLPRLVENLSKVYSPLIGRQIDAFNEILVTSGAYEALYSTILGHVDTGDEVIIIEPYFDCYDFMVKCAGGVPRFIALKPKPQGDDISSADWVLDEAELASLFNNRTKMIIVNTPHNPLGKVFTQRELELIADLCKKHNVLCLSDEVYEWMVYEPKQHIRIASFPGMWERTITVGSAGKTFSVTGWKIGWAYGPADLMRNLQVVHQNCVYTCCTPVEEAVARSFEYELARRDSPDCYFYSLARELRPKRDYLMKILKENGFKPTLPEAGYFVVADWTDLEKKIDLSSELDKYKDYKFTKKFAKEAGVLAIPPSAFYSEAHKHLGETFARFCFIKKDENLELTENLLKEWNSKKK
Summary
Uniprot
H9JEH4
A0A194R9M9
I4DK99
A0A194PPR4
A0A3S2LGC2
A0A212EJW4
+ More
A0A2H1VEV1 A0A2A4JWE0 A0A2A4JWE4 A0A1I8QB36 A0A1I8M5D8 A0A1I8M5F1 T1P7L4 A0A0L0BWG1 A0A1A9X6H3 A0A1B0BNX6 A0A1A9ZCE4 A0A034WA77 A0A034WBU1 A0A0K8UPD0 W8C121 W8BRD0 D3TNC6 A0A0K8UGJ2 A0A1A9UIH9 A0A023ESY6 A0A067RU04 A0A1W7R8M2 A0A182G5N1 A0A2J7Q0K7 A0A336M8U5 A0A1A9WRS4 B4HHW5 B4QTK5 Q17CS8 Q95VY4 A0A1L8E359 B3P4E6 Q8SXC2 A0A1L8E2G2 A0A232FEA9 A0A182JNB8 B3MTT3 A0A182UTQ1 A0A2A3EAC7 V9IJN5 A0A182UIE8 A0A1W4WD68 A0A182X5M5 A0A286T8A1 A0A182KL37 A0A182HWC4 B4GMB8 B0WL67 B4PTF3 Q7QHE7 A0A1S4H6Y0 Q295B7 A0A182N2B6 A0A182JIJ1 A0A1B6H6H8 J3JZA5 A0A0M4ERB4 A0A1Q3FG75 B4LYB3 A0A0Q9WEL4 A0A0Q9WFZ1 A0A139WA99 B4KB46 A0A0Q9WM31 A0A0K8TPA3 A0A1Y1NFN0 A0A286T8E9 E2BA79 A0A182NZM3 A0A0C9RM02 A0A182QZH0 A0A0C9REP2 A0A026WRU1 A0A182WDI7 A0A1B6C996 A0A182RTD1 A0A411G6J5 A0A182FKK8 A0A182LX70 A0A3B0KIX5 A0A2M3Z2M3 A0A2M3Z2K7 A0A2M3ZGA7 A0A2M4BMR4 A0A182YB24 A0A2M3Z2Q7 A0A2M4BN44 A0A084VHT5 A0A182SZM6 B4JG29 A0A2M4A4F3 W5J4G4 A0A151IAF6 E2AHP8
A0A2H1VEV1 A0A2A4JWE0 A0A2A4JWE4 A0A1I8QB36 A0A1I8M5D8 A0A1I8M5F1 T1P7L4 A0A0L0BWG1 A0A1A9X6H3 A0A1B0BNX6 A0A1A9ZCE4 A0A034WA77 A0A034WBU1 A0A0K8UPD0 W8C121 W8BRD0 D3TNC6 A0A0K8UGJ2 A0A1A9UIH9 A0A023ESY6 A0A067RU04 A0A1W7R8M2 A0A182G5N1 A0A2J7Q0K7 A0A336M8U5 A0A1A9WRS4 B4HHW5 B4QTK5 Q17CS8 Q95VY4 A0A1L8E359 B3P4E6 Q8SXC2 A0A1L8E2G2 A0A232FEA9 A0A182JNB8 B3MTT3 A0A182UTQ1 A0A2A3EAC7 V9IJN5 A0A182UIE8 A0A1W4WD68 A0A182X5M5 A0A286T8A1 A0A182KL37 A0A182HWC4 B4GMB8 B0WL67 B4PTF3 Q7QHE7 A0A1S4H6Y0 Q295B7 A0A182N2B6 A0A182JIJ1 A0A1B6H6H8 J3JZA5 A0A0M4ERB4 A0A1Q3FG75 B4LYB3 A0A0Q9WEL4 A0A0Q9WFZ1 A0A139WA99 B4KB46 A0A0Q9WM31 A0A0K8TPA3 A0A1Y1NFN0 A0A286T8E9 E2BA79 A0A182NZM3 A0A0C9RM02 A0A182QZH0 A0A0C9REP2 A0A026WRU1 A0A182WDI7 A0A1B6C996 A0A182RTD1 A0A411G6J5 A0A182FKK8 A0A182LX70 A0A3B0KIX5 A0A2M3Z2M3 A0A2M3Z2K7 A0A2M3ZGA7 A0A2M4BMR4 A0A182YB24 A0A2M3Z2Q7 A0A2M4BN44 A0A084VHT5 A0A182SZM6 B4JG29 A0A2M4A4F3 W5J4G4 A0A151IAF6 E2AHP8
Pubmed
19121390
26354079
22651552
22118469
25315136
26108605
+ More
25348373 24495485 20353571 24945155 24845553 26483478 17994087 17510324 12110301 11880382 15853804 18186649 29113027 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 18057021 20966253 17550304 12364791 15632085 23185243 22516182 18362917 19820115 26369729 28004739 20798317 24508170 30249741 25244985 24438588 20920257 23761445
25348373 24495485 20353571 24945155 24845553 26483478 17994087 17510324 12110301 11880382 15853804 18186649 29113027 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28648823 18057021 20966253 17550304 12364791 15632085 23185243 22516182 18362917 19820115 26369729 28004739 20798317 24508170 30249741 25244985 24438588 20920257 23761445
EMBL
BABH01003100
BABH01003101
BABH01003102
BABH01003103
BABH01003104
KQ460473
+ More
KPJ14538.1 AK401717 BAM18339.1 KQ459596 KPI95436.1 RSAL01000014 RVE53222.1 AGBW02014392 OWR41764.1 ODYU01002164 SOQ39306.1 NWSH01000550 PCG75710.1 PCG75712.1 KA644581 AFP59210.1 JRES01001243 KNC24356.1 JXJN01017657 GAKP01007349 GAKP01007348 JAC51604.1 GAKP01007347 JAC51605.1 GDHF01023966 JAI28348.1 GAMC01007149 JAB99406.1 GAMC01007150 GAMC01007148 GAMC01007147 JAB99408.1 EZ422928 ADD19204.1 GDHF01026621 JAI25693.1 GAPW01001191 JAC12407.1 KK852463 KDR23324.1 GEHC01000161 JAV47484.1 JXUM01146375 KQ570161 KXJ68439.1 NEVH01019967 PNF22096.1 UFQS01000562 UFQT01000562 UFQT01001505 SSX04967.1 SSX25329.1 SSX30867.1 CH480815 EDW42547.1 CM000364 EDX13310.1 CH477304 EAT44194.1 AF395204 AAK97625.1 GFDF01001159 JAV12925.1 CH954181 EDV49461.1 AE014297 AY094691 BT083433 AAF54707.2 AAF54709.2 AAM11044.1 ACQ99838.1 GFDF01001220 JAV12864.1 NNAY01000367 OXU28850.1 CH902623 EDV30214.2 KPU72739.1 KZ288312 PBC28424.1 JR047850 AEY60524.1 LC260184 BBA45756.1 APCN01001484 CH479185 EDW37992.1 DS231980 EDS30240.1 CM000160 EDW97652.1 KRK03881.1 KRK03882.1 AAAB01008816 EAA05134.4 CM000070 EAL28795.2 KRT01066.1 KRT01067.1 GECU01037434 GECU01029077 GECU01019781 JAS70272.1 JAS78629.1 JAS87925.1 BT128586 AEE63543.1 CP012526 ALC47022.1 GFDL01008500 JAV26545.1 CH940650 EDW66909.2 KRF83000.1 KRF82998.1 KQ971410 KYB24841.1 CH933806 EDW15750.2 KRG01710.1 KRG01711.1 KRF82999.1 GDAI01001424 JAI16179.1 GEZM01003558 JAV96734.1 LC260185 BBA45757.1 GL446678 EFN87421.1 GBYB01008006 JAG77773.1 AXCN02001729 GBYB01006789 GBYB01006790 JAG76556.1 JAG76557.1 KK107119 QOIP01000009 EZA58648.1 RLU18712.1 GEDC01027267 GEDC01019560 JAS10031.1 JAS17738.1 MH365753 QBB01562.1 AXCM01001359 OUUW01000014 SPP88480.1 GGFM01002015 MBW22766.1 GGFM01002011 MBW22762.1 GGFM01006835 MBW27586.1 GGFJ01005204 MBW54345.1 GGFM01002022 MBW22773.1 GGFJ01005242 MBW54383.1 ATLV01013235 KE524847 KFB37529.1 CH916369 EDV93596.1 GGFK01002289 MBW35610.1 ADMH02002090 ETN59357.1 KQ978221 KYM96289.1 GL439579 EFN67028.1
KPJ14538.1 AK401717 BAM18339.1 KQ459596 KPI95436.1 RSAL01000014 RVE53222.1 AGBW02014392 OWR41764.1 ODYU01002164 SOQ39306.1 NWSH01000550 PCG75710.1 PCG75712.1 KA644581 AFP59210.1 JRES01001243 KNC24356.1 JXJN01017657 GAKP01007349 GAKP01007348 JAC51604.1 GAKP01007347 JAC51605.1 GDHF01023966 JAI28348.1 GAMC01007149 JAB99406.1 GAMC01007150 GAMC01007148 GAMC01007147 JAB99408.1 EZ422928 ADD19204.1 GDHF01026621 JAI25693.1 GAPW01001191 JAC12407.1 KK852463 KDR23324.1 GEHC01000161 JAV47484.1 JXUM01146375 KQ570161 KXJ68439.1 NEVH01019967 PNF22096.1 UFQS01000562 UFQT01000562 UFQT01001505 SSX04967.1 SSX25329.1 SSX30867.1 CH480815 EDW42547.1 CM000364 EDX13310.1 CH477304 EAT44194.1 AF395204 AAK97625.1 GFDF01001159 JAV12925.1 CH954181 EDV49461.1 AE014297 AY094691 BT083433 AAF54707.2 AAF54709.2 AAM11044.1 ACQ99838.1 GFDF01001220 JAV12864.1 NNAY01000367 OXU28850.1 CH902623 EDV30214.2 KPU72739.1 KZ288312 PBC28424.1 JR047850 AEY60524.1 LC260184 BBA45756.1 APCN01001484 CH479185 EDW37992.1 DS231980 EDS30240.1 CM000160 EDW97652.1 KRK03881.1 KRK03882.1 AAAB01008816 EAA05134.4 CM000070 EAL28795.2 KRT01066.1 KRT01067.1 GECU01037434 GECU01029077 GECU01019781 JAS70272.1 JAS78629.1 JAS87925.1 BT128586 AEE63543.1 CP012526 ALC47022.1 GFDL01008500 JAV26545.1 CH940650 EDW66909.2 KRF83000.1 KRF82998.1 KQ971410 KYB24841.1 CH933806 EDW15750.2 KRG01710.1 KRG01711.1 KRF82999.1 GDAI01001424 JAI16179.1 GEZM01003558 JAV96734.1 LC260185 BBA45757.1 GL446678 EFN87421.1 GBYB01008006 JAG77773.1 AXCN02001729 GBYB01006789 GBYB01006790 JAG76556.1 JAG76557.1 KK107119 QOIP01000009 EZA58648.1 RLU18712.1 GEDC01027267 GEDC01019560 JAS10031.1 JAS17738.1 MH365753 QBB01562.1 AXCM01001359 OUUW01000014 SPP88480.1 GGFM01002015 MBW22766.1 GGFM01002011 MBW22762.1 GGFM01006835 MBW27586.1 GGFJ01005204 MBW54345.1 GGFM01002022 MBW22773.1 GGFJ01005242 MBW54383.1 ATLV01013235 KE524847 KFB37529.1 CH916369 EDV93596.1 GGFK01002289 MBW35610.1 ADMH02002090 ETN59357.1 KQ978221 KYM96289.1 GL439579 EFN67028.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000218220
+ More
UP000095300 UP000095301 UP000037069 UP000092443 UP000092460 UP000092445 UP000078200 UP000027135 UP000069940 UP000249989 UP000235965 UP000091820 UP000001292 UP000000304 UP000008820 UP000008711 UP000000803 UP000215335 UP000075881 UP000007801 UP000075903 UP000242457 UP000075902 UP000192221 UP000076407 UP000075882 UP000075840 UP000008744 UP000002320 UP000002282 UP000007062 UP000001819 UP000075884 UP000075880 UP000092553 UP000008792 UP000007266 UP000009192 UP000008237 UP000075885 UP000075886 UP000053097 UP000279307 UP000075920 UP000075900 UP000069272 UP000075883 UP000268350 UP000076408 UP000030765 UP000075901 UP000001070 UP000000673 UP000078542 UP000000311
UP000095300 UP000095301 UP000037069 UP000092443 UP000092460 UP000092445 UP000078200 UP000027135 UP000069940 UP000249989 UP000235965 UP000091820 UP000001292 UP000000304 UP000008820 UP000008711 UP000000803 UP000215335 UP000075881 UP000007801 UP000075903 UP000242457 UP000075902 UP000192221 UP000076407 UP000075882 UP000075840 UP000008744 UP000002320 UP000002282 UP000007062 UP000001819 UP000075884 UP000075880 UP000092553 UP000008792 UP000007266 UP000009192 UP000008237 UP000075885 UP000075886 UP000053097 UP000279307 UP000075920 UP000075900 UP000069272 UP000075883 UP000268350 UP000076408 UP000030765 UP000075901 UP000001070 UP000000673 UP000078542 UP000000311
PRIDE
Pfam
PF00155 Aminotran_1_2
Interpro
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
H9JEH4
A0A194R9M9
I4DK99
A0A194PPR4
A0A3S2LGC2
A0A212EJW4
+ More
A0A2H1VEV1 A0A2A4JWE0 A0A2A4JWE4 A0A1I8QB36 A0A1I8M5D8 A0A1I8M5F1 T1P7L4 A0A0L0BWG1 A0A1A9X6H3 A0A1B0BNX6 A0A1A9ZCE4 A0A034WA77 A0A034WBU1 A0A0K8UPD0 W8C121 W8BRD0 D3TNC6 A0A0K8UGJ2 A0A1A9UIH9 A0A023ESY6 A0A067RU04 A0A1W7R8M2 A0A182G5N1 A0A2J7Q0K7 A0A336M8U5 A0A1A9WRS4 B4HHW5 B4QTK5 Q17CS8 Q95VY4 A0A1L8E359 B3P4E6 Q8SXC2 A0A1L8E2G2 A0A232FEA9 A0A182JNB8 B3MTT3 A0A182UTQ1 A0A2A3EAC7 V9IJN5 A0A182UIE8 A0A1W4WD68 A0A182X5M5 A0A286T8A1 A0A182KL37 A0A182HWC4 B4GMB8 B0WL67 B4PTF3 Q7QHE7 A0A1S4H6Y0 Q295B7 A0A182N2B6 A0A182JIJ1 A0A1B6H6H8 J3JZA5 A0A0M4ERB4 A0A1Q3FG75 B4LYB3 A0A0Q9WEL4 A0A0Q9WFZ1 A0A139WA99 B4KB46 A0A0Q9WM31 A0A0K8TPA3 A0A1Y1NFN0 A0A286T8E9 E2BA79 A0A182NZM3 A0A0C9RM02 A0A182QZH0 A0A0C9REP2 A0A026WRU1 A0A182WDI7 A0A1B6C996 A0A182RTD1 A0A411G6J5 A0A182FKK8 A0A182LX70 A0A3B0KIX5 A0A2M3Z2M3 A0A2M3Z2K7 A0A2M3ZGA7 A0A2M4BMR4 A0A182YB24 A0A2M3Z2Q7 A0A2M4BN44 A0A084VHT5 A0A182SZM6 B4JG29 A0A2M4A4F3 W5J4G4 A0A151IAF6 E2AHP8
A0A2H1VEV1 A0A2A4JWE0 A0A2A4JWE4 A0A1I8QB36 A0A1I8M5D8 A0A1I8M5F1 T1P7L4 A0A0L0BWG1 A0A1A9X6H3 A0A1B0BNX6 A0A1A9ZCE4 A0A034WA77 A0A034WBU1 A0A0K8UPD0 W8C121 W8BRD0 D3TNC6 A0A0K8UGJ2 A0A1A9UIH9 A0A023ESY6 A0A067RU04 A0A1W7R8M2 A0A182G5N1 A0A2J7Q0K7 A0A336M8U5 A0A1A9WRS4 B4HHW5 B4QTK5 Q17CS8 Q95VY4 A0A1L8E359 B3P4E6 Q8SXC2 A0A1L8E2G2 A0A232FEA9 A0A182JNB8 B3MTT3 A0A182UTQ1 A0A2A3EAC7 V9IJN5 A0A182UIE8 A0A1W4WD68 A0A182X5M5 A0A286T8A1 A0A182KL37 A0A182HWC4 B4GMB8 B0WL67 B4PTF3 Q7QHE7 A0A1S4H6Y0 Q295B7 A0A182N2B6 A0A182JIJ1 A0A1B6H6H8 J3JZA5 A0A0M4ERB4 A0A1Q3FG75 B4LYB3 A0A0Q9WEL4 A0A0Q9WFZ1 A0A139WA99 B4KB46 A0A0Q9WM31 A0A0K8TPA3 A0A1Y1NFN0 A0A286T8E9 E2BA79 A0A182NZM3 A0A0C9RM02 A0A182QZH0 A0A0C9REP2 A0A026WRU1 A0A182WDI7 A0A1B6C996 A0A182RTD1 A0A411G6J5 A0A182FKK8 A0A182LX70 A0A3B0KIX5 A0A2M3Z2M3 A0A2M3Z2K7 A0A2M3ZGA7 A0A2M4BMR4 A0A182YB24 A0A2M3Z2Q7 A0A2M4BN44 A0A084VHT5 A0A182SZM6 B4JG29 A0A2M4A4F3 W5J4G4 A0A151IAF6 E2AHP8
PDB
2R5E
E-value=1.05829e-156,
Score=1420
Ontologies
PATHWAY
GO
PANTHER
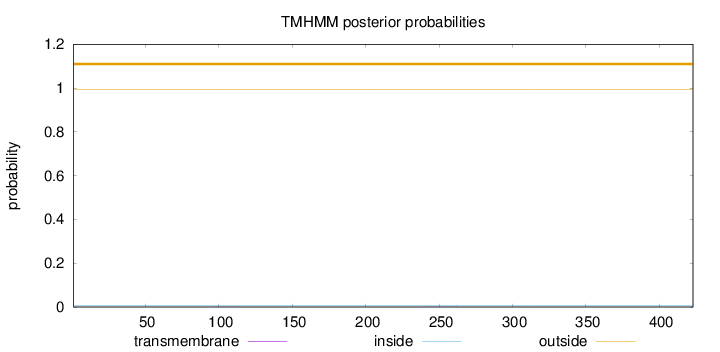
Topology
Length:
423
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00953
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00764
outside
1 - 423
Population Genetic Test Statistics
Pi
222.532427
Theta
150.170827
Tajima's D
2.245783
CLR
0.435576
CSRT
0.915904204789761
Interpretation
Uncertain
