Gene
KWMTBOMO08704
Pre Gene Modal
BGIBMGA007923
Annotation
PREDICTED:_histone-lysine_N-methyltransferase_ASHR1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.386 Nuclear Reliability : 1.52
Sequence
CDS
ATGAGGAGCAAATTCAGAAATACAACTTCCGAAATAAAACCTGGAGATCTTTTGTTGTCAGAGCAACCATTCGCTTATGTATTATCGTCCAAAGAACAAGGAAACCGATGTGATAATTGCTTAGAAAAAGTCAAACTCCTAAAATGTTCAGGATGTCAATTTGTTTATTACTGTGGAAGATCTTGTCAGAAAGATGCATGGGGAGATCACAAGTGGGAGTGCCCTAATCTCAAACGAGTCGCGCCAAAAGTAATCCCTGATGGCGCACGAATGCTGGCAAAGATCATCAACAGGTTGAACCGGGGCGATGGACACTCGTTTAGGTCTTTTTATTCGGCTACCTCATTTAGAATGTGGAAGGATCTCATGTCACATTATCCTAATTTGAAAGCAGACAAGAAAAGGATGGATCATTTCACTTCATTATGCGTTGTGCTGTATGAATTTTTAAAAGACATTTCTCTACCAAACACAGTTGAGCTAATGGGGCTTTATGGACGGATGGTGATAAACAGCTTCATGATCTTAGATACAGACATGAATTCGATTGGTACCGGTATTTATTTGGCTTCCTCCATAGTGGACCACAGTTGCGAACCAAACGCTGTGGTCACGTTTGACGGAAAGACAATCAACATCAGAGCGATACAAAACATGTCTTACTTAGACTGGAATCAGATACGCATATCTTATATCGACATGATGAAAACACCGTATGAAAGACAAAATGAGTTGTTGAGCAGCTATTACTTCTTGTGTGGATGTCACAGGTGCATCGATGGAAATCAAGTTAAATATGTTCACGCCGCCACATGCTTAAACGAAAACTGTGAAGCTCCCGTAGAGATTACCTGGAATGAAAACTGTCAGCTCGTCTTACGACCCGAGACTGAGGGGTCAACGCAAAATGGCGTATCGAACGGGATTAAAAATGGCGACTTTCAAAAACAAAATGGCGGTTGCGACGAGGAGGAAATTGATAACGACGAATTCGTTCACTGCAAAGAATGTGGAACTAAATATACGGAGAAACACGTGGAAGCTTTCATTAAAATCATGGAATTCACTAAACAACATTTAAAGAATATGGAGGGTAGTTCTGTGGCATATGTGGACGTTTGCAACTACTGCCTGGACCGTCACGAGGGCGTGCTGCATCCGTTGAACGTGATGCACGCTCAGACTCTGGACCACGCCTTCGAGGCTCACGTACAGATGCAGCTGTGGGAGAAGGCGTGCGTATACGCCGAAAGACTCATTCCGTGTTTCAGGTTTTATCACGGCGACAAGAATCCTCTATTGGGCCTGTTATATCTGAAGTACGGTAAGATCCTGCTGTACAAGATGGATTTGGAAAAGGCCGTTGCGCAACTGAAACTCGCTGAGAAAATCATAAAGACAACTCACGGAGAATCGCATCCTCTTTACAGAGAACAATTGGTGCCTTTGCTTCAACAAGCCATGGCGGAGGCGGATTGA
Protein
MRSKFRNTTSEIKPGDLLLSEQPFAYVLSSKEQGNRCDNCLEKVKLLKCSGCQFVYYCGRSCQKDAWGDHKWECPNLKRVAPKVIPDGARMLAKIINRLNRGDGHSFRSFYSATSFRMWKDLMSHYPNLKADKKRMDHFTSLCVVLYEFLKDISLPNTVELMGLYGRMVINSFMILDTDMNSIGTGIYLASSIVDHSCEPNAVVTFDGKTINIRAIQNMSYLDWNQIRISYIDMMKTPYERQNELLSSYYFLCGCHRCIDGNQVKYVHAATCLNENCEAPVEITWNENCQLVLRPETEGSTQNGVSNGIKNGDFQKQNGGCDEEEIDNDEFVHCKECGTKYTEKHVEAFIKIMEFTKQHLKNMEGSSVAYVDVCNYCLDRHEGVLHPLNVMHAQTLDHAFEAHVQMQLWEKACVYAERLIPCFRFYHGDKNPLLGLLYLKYGKILLYKMDLEKAVAQLKLAEKIIKTTHGESHPLYREQLVPLLQQAMAEAD
Summary
Uniprot
A0A2H1V1Z6
A0A2A4JWD6
A0A437BIE5
A0A1E1WS13
A0A194PQ37
A0A212EMH4
+ More
A0A2J7QV68 A0A067RS12 D6WD02 A0A1B6DNR1 A0A0L7R4Z8 A0A1Y1LKX3 A0A1Y1LR32 W8B957 A0A0L0C874 A0A088A5F5 T1P9V7 A0A1I8MCF6 A0A1B0DRH3 A0A1B0FJ24 A0A1I8MCF8 B3MYX1 A0A154PCX6 A0A1A9ZEL1 A0A2A3EGD6 A0A1A9WZC6 A0A034VWS3 B4MA84 A0A0R3P6J9 Q29GT9 A0A0A1WS40 A0A2J7QV83 Q9W4X8 A0A026W1V0 B4L709 B3P934 A0A3B0JYS0 A0A3L8DL22 A0A195CI34 A0A2P8XLB6 A0A336MBH9 B4I9X0 A0A1B6JWE4 A0A1B6M851 A0A1W4V8U3 A0A1I8NU05 A0A1I8NTZ9 B4Q0Y8 A0A1I8NU13 A0A1B0BDY0 A0A1A9XBS6 B4JX66 A0A1B6KCD1 A0A1A9VCM8 A0A195B778 B4JX63 A0A1B6GW03 A0A158NG82 B4N1L1 A0A0M4ET43 A0A151J5J6 J9JW25 A0A195FFL0 A0A0C9PKW1 A0A0C9PZA1 A0A2S2Q7N1 A0A2S2NWC3 A0A182Y0G1 Q7PWI2 A0A1S4GZR2 A0A182XNV5 A0A182JWV8 A0A182VMW1 A0A232ER57 A0A182TYS9 A0A310SPX0 B4GUY5 A0A182LMN6 A0A182PMG0 B0XCL1 A0A182LU15 A0A1Q3FDL6 A0A182I7G0 A0A182WGA2 A0A0K8TK56 A0A2R7VYB4 A0A084WQG6 Q17EQ8 A0A1J1J705 U4U294 A0A182RJY7 A0A182NUR0 N6T2G0 A0A1S4F5C5 F4WND1 A0A2M4BMB5 A0A182F520 A0A182IJI7
A0A2J7QV68 A0A067RS12 D6WD02 A0A1B6DNR1 A0A0L7R4Z8 A0A1Y1LKX3 A0A1Y1LR32 W8B957 A0A0L0C874 A0A088A5F5 T1P9V7 A0A1I8MCF6 A0A1B0DRH3 A0A1B0FJ24 A0A1I8MCF8 B3MYX1 A0A154PCX6 A0A1A9ZEL1 A0A2A3EGD6 A0A1A9WZC6 A0A034VWS3 B4MA84 A0A0R3P6J9 Q29GT9 A0A0A1WS40 A0A2J7QV83 Q9W4X8 A0A026W1V0 B4L709 B3P934 A0A3B0JYS0 A0A3L8DL22 A0A195CI34 A0A2P8XLB6 A0A336MBH9 B4I9X0 A0A1B6JWE4 A0A1B6M851 A0A1W4V8U3 A0A1I8NU05 A0A1I8NTZ9 B4Q0Y8 A0A1I8NU13 A0A1B0BDY0 A0A1A9XBS6 B4JX66 A0A1B6KCD1 A0A1A9VCM8 A0A195B778 B4JX63 A0A1B6GW03 A0A158NG82 B4N1L1 A0A0M4ET43 A0A151J5J6 J9JW25 A0A195FFL0 A0A0C9PKW1 A0A0C9PZA1 A0A2S2Q7N1 A0A2S2NWC3 A0A182Y0G1 Q7PWI2 A0A1S4GZR2 A0A182XNV5 A0A182JWV8 A0A182VMW1 A0A232ER57 A0A182TYS9 A0A310SPX0 B4GUY5 A0A182LMN6 A0A182PMG0 B0XCL1 A0A182LU15 A0A1Q3FDL6 A0A182I7G0 A0A182WGA2 A0A0K8TK56 A0A2R7VYB4 A0A084WQG6 Q17EQ8 A0A1J1J705 U4U294 A0A182RJY7 A0A182NUR0 N6T2G0 A0A1S4F5C5 F4WND1 A0A2M4BMB5 A0A182F520 A0A182IJI7
Pubmed
26354079
22118469
24845553
18362917
19820115
28004739
+ More
24495485 26108605 25315136 17994087 25348373 15632085 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741 29403074 17550304 21347285 25244985 12364791 28648823 20966253 26369729 24438588 17510324 23537049 21719571
24495485 26108605 25315136 17994087 25348373 15632085 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741 29403074 17550304 21347285 25244985 12364791 28648823 20966253 26369729 24438588 17510324 23537049 21719571
EMBL
ODYU01000320
SOQ34851.1
NWSH01000550
PCG75702.1
RSAL01000051
RVE50206.1
+ More
GDQN01001276 JAT89778.1 KQ459596 KPI95427.1 AGBW02013840 OWR42677.1 NEVH01010478 PNF32480.1 KK852460 KDR23505.1 KQ971311 EEZ98820.1 GEDC01009991 GEDC01000606 JAS27307.1 JAS36692.1 KQ414654 KOC65960.1 GEZM01054686 JAV73488.1 GEZM01054687 JAV73487.1 GAMC01008765 JAB97790.1 JRES01000767 KNC28461.1 KA645546 AFP60175.1 AJVK01020029 CCAG010017454 CH902632 EDV32815.1 KQ434874 KZC09745.1 KZ288256 PBC30544.1 GAKP01012401 GAKP01012400 GAKP01012399 GAKP01012396 JAC46552.1 CH940655 EDW66143.1 CH379064 KRT06774.1 EAL32020.1 GBXI01012620 JAD01672.1 PNF32481.1 AE014298 BT060437 AL121804 AAF45795.2 ACN22202.1 CAB58065.1 KK107487 EZA50042.1 CH933812 EDW06155.1 CH954183 EDV45639.1 OUUW01000003 SPP77871.1 QOIP01000006 RLU21117.1 KQ977721 KYN00400.1 PYGN01001793 PSN32785.1 UFQS01000653 UFQT01000653 SSX05804.1 SSX26163.1 CH480825 EDW44001.1 GECU01004196 JAT03511.1 GEBQ01007896 JAT32081.1 CM000162 EDX01355.1 JXJN01012727 CH916376 EDV95342.1 GEBQ01030858 JAT09119.1 KQ976574 KYM80351.1 EDV95339.1 GECZ01007614 GECZ01003192 JAS62155.1 JAS66577.1 ADTU01015041 CH963925 EDW78250.1 CP012528 ALC49602.1 KQ980017 KYN18233.1 ABLF02039328 KQ981625 KYN39012.1 GBYB01001638 JAG71405.1 GBYB01006848 JAG76615.1 GGMS01004367 MBY73570.1 GGMR01008669 MBY21288.1 AAAB01008984 EAA14812.4 NNAY01002670 OXU20812.1 KQ761783 OAD56993.1 CH479192 EDW26522.1 DS232704 EDS44907.1 AXCM01002875 GFDL01009389 JAV25656.1 APCN01003394 GDAI01002996 JAI14607.1 KK854135 PTY11911.1 ATLV01025608 KE525396 KFB52460.1 CH477279 EAT45044.1 CVRI01000072 CRL07564.1 KB631924 ERL87192.1 APGK01055127 KB741259 ENN71728.1 GL888236 EGI64301.1 GGFJ01005031 MBW54172.1
GDQN01001276 JAT89778.1 KQ459596 KPI95427.1 AGBW02013840 OWR42677.1 NEVH01010478 PNF32480.1 KK852460 KDR23505.1 KQ971311 EEZ98820.1 GEDC01009991 GEDC01000606 JAS27307.1 JAS36692.1 KQ414654 KOC65960.1 GEZM01054686 JAV73488.1 GEZM01054687 JAV73487.1 GAMC01008765 JAB97790.1 JRES01000767 KNC28461.1 KA645546 AFP60175.1 AJVK01020029 CCAG010017454 CH902632 EDV32815.1 KQ434874 KZC09745.1 KZ288256 PBC30544.1 GAKP01012401 GAKP01012400 GAKP01012399 GAKP01012396 JAC46552.1 CH940655 EDW66143.1 CH379064 KRT06774.1 EAL32020.1 GBXI01012620 JAD01672.1 PNF32481.1 AE014298 BT060437 AL121804 AAF45795.2 ACN22202.1 CAB58065.1 KK107487 EZA50042.1 CH933812 EDW06155.1 CH954183 EDV45639.1 OUUW01000003 SPP77871.1 QOIP01000006 RLU21117.1 KQ977721 KYN00400.1 PYGN01001793 PSN32785.1 UFQS01000653 UFQT01000653 SSX05804.1 SSX26163.1 CH480825 EDW44001.1 GECU01004196 JAT03511.1 GEBQ01007896 JAT32081.1 CM000162 EDX01355.1 JXJN01012727 CH916376 EDV95342.1 GEBQ01030858 JAT09119.1 KQ976574 KYM80351.1 EDV95339.1 GECZ01007614 GECZ01003192 JAS62155.1 JAS66577.1 ADTU01015041 CH963925 EDW78250.1 CP012528 ALC49602.1 KQ980017 KYN18233.1 ABLF02039328 KQ981625 KYN39012.1 GBYB01001638 JAG71405.1 GBYB01006848 JAG76615.1 GGMS01004367 MBY73570.1 GGMR01008669 MBY21288.1 AAAB01008984 EAA14812.4 NNAY01002670 OXU20812.1 KQ761783 OAD56993.1 CH479192 EDW26522.1 DS232704 EDS44907.1 AXCM01002875 GFDL01009389 JAV25656.1 APCN01003394 GDAI01002996 JAI14607.1 KK854135 PTY11911.1 ATLV01025608 KE525396 KFB52460.1 CH477279 EAT45044.1 CVRI01000072 CRL07564.1 KB631924 ERL87192.1 APGK01055127 KB741259 ENN71728.1 GL888236 EGI64301.1 GGFJ01005031 MBW54172.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007151
UP000235965
UP000027135
+ More
UP000007266 UP000053825 UP000037069 UP000005203 UP000095301 UP000092462 UP000092444 UP000007801 UP000076502 UP000092445 UP000242457 UP000091820 UP000008792 UP000001819 UP000000803 UP000053097 UP000009192 UP000008711 UP000268350 UP000279307 UP000078542 UP000245037 UP000001292 UP000192221 UP000095300 UP000002282 UP000092460 UP000092443 UP000001070 UP000078200 UP000078540 UP000005205 UP000007798 UP000092553 UP000078492 UP000007819 UP000078541 UP000076408 UP000007062 UP000076407 UP000075881 UP000075903 UP000215335 UP000075902 UP000008744 UP000075882 UP000075885 UP000002320 UP000075883 UP000075840 UP000075920 UP000030765 UP000008820 UP000183832 UP000030742 UP000075900 UP000075884 UP000019118 UP000007755 UP000069272 UP000075880
UP000007266 UP000053825 UP000037069 UP000005203 UP000095301 UP000092462 UP000092444 UP000007801 UP000076502 UP000092445 UP000242457 UP000091820 UP000008792 UP000001819 UP000000803 UP000053097 UP000009192 UP000008711 UP000268350 UP000279307 UP000078542 UP000245037 UP000001292 UP000192221 UP000095300 UP000002282 UP000092460 UP000092443 UP000001070 UP000078200 UP000078540 UP000005205 UP000007798 UP000092553 UP000078492 UP000007819 UP000078541 UP000076408 UP000007062 UP000076407 UP000075881 UP000075903 UP000215335 UP000075902 UP000008744 UP000075882 UP000075885 UP000002320 UP000075883 UP000075840 UP000075920 UP000030765 UP000008820 UP000183832 UP000030742 UP000075900 UP000075884 UP000019118 UP000007755 UP000069272 UP000075880
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A2H1V1Z6
A0A2A4JWD6
A0A437BIE5
A0A1E1WS13
A0A194PQ37
A0A212EMH4
+ More
A0A2J7QV68 A0A067RS12 D6WD02 A0A1B6DNR1 A0A0L7R4Z8 A0A1Y1LKX3 A0A1Y1LR32 W8B957 A0A0L0C874 A0A088A5F5 T1P9V7 A0A1I8MCF6 A0A1B0DRH3 A0A1B0FJ24 A0A1I8MCF8 B3MYX1 A0A154PCX6 A0A1A9ZEL1 A0A2A3EGD6 A0A1A9WZC6 A0A034VWS3 B4MA84 A0A0R3P6J9 Q29GT9 A0A0A1WS40 A0A2J7QV83 Q9W4X8 A0A026W1V0 B4L709 B3P934 A0A3B0JYS0 A0A3L8DL22 A0A195CI34 A0A2P8XLB6 A0A336MBH9 B4I9X0 A0A1B6JWE4 A0A1B6M851 A0A1W4V8U3 A0A1I8NU05 A0A1I8NTZ9 B4Q0Y8 A0A1I8NU13 A0A1B0BDY0 A0A1A9XBS6 B4JX66 A0A1B6KCD1 A0A1A9VCM8 A0A195B778 B4JX63 A0A1B6GW03 A0A158NG82 B4N1L1 A0A0M4ET43 A0A151J5J6 J9JW25 A0A195FFL0 A0A0C9PKW1 A0A0C9PZA1 A0A2S2Q7N1 A0A2S2NWC3 A0A182Y0G1 Q7PWI2 A0A1S4GZR2 A0A182XNV5 A0A182JWV8 A0A182VMW1 A0A232ER57 A0A182TYS9 A0A310SPX0 B4GUY5 A0A182LMN6 A0A182PMG0 B0XCL1 A0A182LU15 A0A1Q3FDL6 A0A182I7G0 A0A182WGA2 A0A0K8TK56 A0A2R7VYB4 A0A084WQG6 Q17EQ8 A0A1J1J705 U4U294 A0A182RJY7 A0A182NUR0 N6T2G0 A0A1S4F5C5 F4WND1 A0A2M4BMB5 A0A182F520 A0A182IJI7
A0A2J7QV68 A0A067RS12 D6WD02 A0A1B6DNR1 A0A0L7R4Z8 A0A1Y1LKX3 A0A1Y1LR32 W8B957 A0A0L0C874 A0A088A5F5 T1P9V7 A0A1I8MCF6 A0A1B0DRH3 A0A1B0FJ24 A0A1I8MCF8 B3MYX1 A0A154PCX6 A0A1A9ZEL1 A0A2A3EGD6 A0A1A9WZC6 A0A034VWS3 B4MA84 A0A0R3P6J9 Q29GT9 A0A0A1WS40 A0A2J7QV83 Q9W4X8 A0A026W1V0 B4L709 B3P934 A0A3B0JYS0 A0A3L8DL22 A0A195CI34 A0A2P8XLB6 A0A336MBH9 B4I9X0 A0A1B6JWE4 A0A1B6M851 A0A1W4V8U3 A0A1I8NU05 A0A1I8NTZ9 B4Q0Y8 A0A1I8NU13 A0A1B0BDY0 A0A1A9XBS6 B4JX66 A0A1B6KCD1 A0A1A9VCM8 A0A195B778 B4JX63 A0A1B6GW03 A0A158NG82 B4N1L1 A0A0M4ET43 A0A151J5J6 J9JW25 A0A195FFL0 A0A0C9PKW1 A0A0C9PZA1 A0A2S2Q7N1 A0A2S2NWC3 A0A182Y0G1 Q7PWI2 A0A1S4GZR2 A0A182XNV5 A0A182JWV8 A0A182VMW1 A0A232ER57 A0A182TYS9 A0A310SPX0 B4GUY5 A0A182LMN6 A0A182PMG0 B0XCL1 A0A182LU15 A0A1Q3FDL6 A0A182I7G0 A0A182WGA2 A0A0K8TK56 A0A2R7VYB4 A0A084WQG6 Q17EQ8 A0A1J1J705 U4U294 A0A182RJY7 A0A182NUR0 N6T2G0 A0A1S4F5C5 F4WND1 A0A2M4BMB5 A0A182F520 A0A182IJI7
PDB
5HQ8
E-value=3.98268e-39,
Score=406
Ontologies
Topology
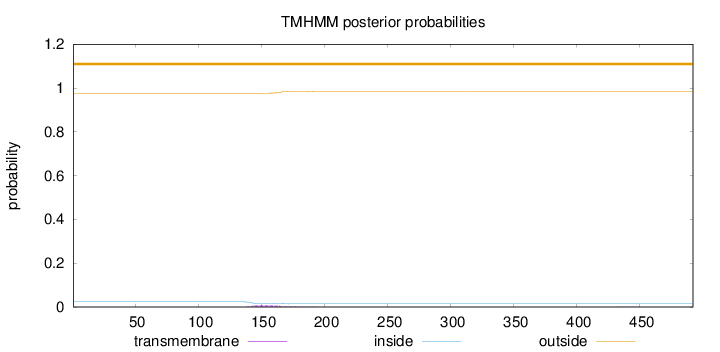
Length:
492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21173
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02371
outside
1 - 492
Population Genetic Test Statistics
Pi
235.301361
Theta
168.27169
Tajima's D
1.130023
CLR
0.000003
CSRT
0.690565471726414
Interpretation
Uncertain
