Gene
KWMTBOMO08703
Pre Gene Modal
BGIBMGA007924
Annotation
PREDICTED:_fizzy-related_protein_homolog_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 2.008 Nuclear Reliability : 2.105
Sequence
CDS
ATGTTCAGTAAACATTATGAGACTCGGTTACTACAGCAGTGCAGTCCTGTGGCAAGGAAACAGTCTGTACTGGTCCAGGCTGAGTTTAACAATGGTGACAGGTTCACACCCTGCAGAGCTGGAAACAATTGGCAGACAGCTTTTCAAGCACTAAGAAGAGGTACGGAAAATCAACCAGCAAAGAAGGCGCGAAACGATGGCGGTGGCAGCGACGGAGCGAGTGGCGGTCGAGACCAGCTCGCTTACAGCTGCCTCCTACAGAACGAACTCCTCGGTGCACAGATAGAGGAAGTCAGGAATCCATCGGCGACGTGTCCGGATGGCAACAAGACTATCGGCACCGTCCAGACGCCTGGCAGATCGTCGCAAGCTTACCAACCGGTCTTCAGATTCAAAGCGCCGATCCACAGAGCAGAGGAAAACATTGATCCTTCGCCGTACAGCCTGTCCCCGATCTCGGCTAAATCCGAGAAGCTTCTCCGGTCCCCAAGGAAGGCGACCCGCAAGATATCGCGGATCCCGTTCAAGGTTCTGGACGCGCCGGAGTTGCAGGACGACTTCTACCTGAACCTGGTCGACTGGTCCGCCCAGAACGTGCTCAGCGTCGGCCTCGGGAGCTGCGTCTACTTGTGGAGCGCCTGCACCAGTCAGGTGACGCGGTTATGTGACCTGTCAACGGAAGGTAACGCGGTGACTTCGGTGGCGTGGAGCGAACGCGGCCACCTCGTCGCCGTCGGGACGCAGAAGGGGCACATCAGCGTGTGGGACGTTGCCGTCAACAAGGAGGTGACGAAGCTCCAAGGTCACATAGCGCGCGTCGGAGCCCTCGCCTGGAACGGTGACATACTCAGCTCAGGGTCCAGGGACCGACACATCCGCCAGCGGGACACCAGAACGCCACCGATACAACTCTCCCGAGTTCTCCAAGGTCACAGACAAGAGGTCTGCGGCCTGAAGTGGTCGCCCGACGGCCAATCGTTGGCTTCAGGAGGCAACGACAACAAGCTCTTCGTTTGGTCGATGCACTCGACGTCGCCGGTGCAGACGTACTCCGCGCACGTGGCGGCCGTGAAGGCGATCGCTTGGTCGCCGCACCAGCACGGGCTGCTCGCGTCCGGCGGCGGCACGGCGGACCGCTGCATACGCTTCTGGAACACGCTCACCTCGCAGCCGATGCAGTGCGTCGACACCGGATCTCAAGTGTGCAACTTGGCTTGGTCGAAACACTCCAGCGAGTTGGTCTCGACTCACGGCTACTCACAGAACCAGATACTGGTGTGGAAATATCCGTCGTTGACGCAGGTGGCGAAACTGACTGGCCATTCCTATCGAGTTCTGTACTTGGCTCTATCACCGGACGGCGAGGCCATCGTCACGGGAGCCGGTGACGAAACGCTTCGGTTCTGGAATGTTTTCTCCAAAACGCCCTCACACAAGGAGAACAAGAGCGTCCTAAACCTATACACCGCACTCAGATAA
Protein
MFSKHYETRLLQQCSPVARKQSVLVQAEFNNGDRFTPCRAGNNWQTAFQALRRGTENQPAKKARNDGGGSDGASGGRDQLAYSCLLQNELLGAQIEEVRNPSATCPDGNKTIGTVQTPGRSSQAYQPVFRFKAPIHRAEENIDPSPYSLSPISAKSEKLLRSPRKATRKISRIPFKVLDAPELQDDFYLNLVDWSAQNVLSVGLGSCVYLWSACTSQVTRLCDLSTEGNAVTSVAWSERGHLVAVGTQKGHISVWDVAVNKEVTKLQGHIARVGALAWNGDILSSGSRDRHIRQRDTRTPPIQLSRVLQGHRQEVCGLKWSPDGQSLASGGNDNKLFVWSMHSTSPVQTYSAHVAAVKAIAWSPHQHGLLASGGGTADRCIRFWNTLTSQPMQCVDTGSQVCNLAWSKHSSELVSTHGYSQNQILVWKYPSLTQVAKLTGHSYRVLYLALSPDGEAIVTGAGDETLRFWNVFSKTPSHKENKSVLNLYTALR
Summary
Cofactor
Mg(2+)
Similarity
Belongs to the PP2C family.
Uniprot
A0A2W1BCE8
A0A3S2NKX6
A0A212EMH9
A0A2A4JW45
A0A194RAM3
H9JEH7
+ More
U5EV22 A0A1I8MAB3 W8ASZ3 A0A1J1HT88 A0A0K8U1X0 Q17CQ5 A0A034VNG0 A0A023ESV6 A0A1Q3G3M5 A0A0A1X5L9 A0A3B0J7B4 Q29G84 B3MS79 B3NU56 A0A1I8NTT0 A0A1W4U5T1 B4L2J6 A0A0M3QZI0 B4NC43 B4R4P5 Q9W4H9 B4Q032 O18402 B4M7U0 B4JX77 A0A336LNN7 A0A182JCA7 A0A340TBF7 A0A182QYL6 A0A232FJF8 Q7PUU4 A0A2J7R4N4 K7ILS6 D6W9Q9 A0A1B6DR46 B4I0W3 A0A195DSV1 F4X2K7 A0A195BT79 A0A195FX20 A0A158NVA2 A0A1B0DPN4 A0A151IBE6 A0A1L8DX39 A0A026WBU6 A0A151WFR8 A0A3L8DNK8 M9PLJ4 E2B214 A0A291S6V5 A0A154PKT1 A0A2A3E3W2 A0A0J7L699 A0A087ZZZ4 E9IVM5 D1LX13 A0A3Q0IXN9 H3A259 E2B8G8 A0A2P8YUD0 A0A0L7R3J1 A0A444TLK5 U6CVQ8 M3XXP4 A0A0C9RIT7 A0A2Y9L6R5 A0A3Q3RQJ7 A0A340XFY9 A0A2Y9M2B6 A0A2K6FS10 A0A0P6J296 A0A091DEE4 A0A286XCI2 A0A341B2D0 A0A3Q7UP78 A0A287D3A8 L5L806 A0A452EMH9 A0A3Q7NB01 A0A2Y9TKN2 A0A2Y9I9T1 A0A337SP30 A0A2U3Y4Y4 W5PSG6 A0A2U3VKG9 A0A212D207 A0A452RRH7 Q5H7B9 Q32L05 A0A250Y1W1 K9ILH8 H0XAM8 A0A383ZNB5 A0A3S3SIC5
U5EV22 A0A1I8MAB3 W8ASZ3 A0A1J1HT88 A0A0K8U1X0 Q17CQ5 A0A034VNG0 A0A023ESV6 A0A1Q3G3M5 A0A0A1X5L9 A0A3B0J7B4 Q29G84 B3MS79 B3NU56 A0A1I8NTT0 A0A1W4U5T1 B4L2J6 A0A0M3QZI0 B4NC43 B4R4P5 Q9W4H9 B4Q032 O18402 B4M7U0 B4JX77 A0A336LNN7 A0A182JCA7 A0A340TBF7 A0A182QYL6 A0A232FJF8 Q7PUU4 A0A2J7R4N4 K7ILS6 D6W9Q9 A0A1B6DR46 B4I0W3 A0A195DSV1 F4X2K7 A0A195BT79 A0A195FX20 A0A158NVA2 A0A1B0DPN4 A0A151IBE6 A0A1L8DX39 A0A026WBU6 A0A151WFR8 A0A3L8DNK8 M9PLJ4 E2B214 A0A291S6V5 A0A154PKT1 A0A2A3E3W2 A0A0J7L699 A0A087ZZZ4 E9IVM5 D1LX13 A0A3Q0IXN9 H3A259 E2B8G8 A0A2P8YUD0 A0A0L7R3J1 A0A444TLK5 U6CVQ8 M3XXP4 A0A0C9RIT7 A0A2Y9L6R5 A0A3Q3RQJ7 A0A340XFY9 A0A2Y9M2B6 A0A2K6FS10 A0A0P6J296 A0A091DEE4 A0A286XCI2 A0A341B2D0 A0A3Q7UP78 A0A287D3A8 L5L806 A0A452EMH9 A0A3Q7NB01 A0A2Y9TKN2 A0A2Y9I9T1 A0A337SP30 A0A2U3Y4Y4 W5PSG6 A0A2U3VKG9 A0A212D207 A0A452RRH7 Q5H7B9 Q32L05 A0A250Y1W1 K9ILH8 H0XAM8 A0A383ZNB5 A0A3S3SIC5
Pubmed
28756777
22118469
26354079
19121390
25315136
24495485
+ More
17510324 25348373 24945155 25830018 15632085 17994087 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 9288747 28648823 12364791 14747013 17210077 20075255 18362917 19820115 21719571 21347285 24508170 30249741 25581109 20798317 28992199 21282665 9215903 29403074 21993624 23258410 17975172 20809919 30723633 19393038 28087693
17510324 25348373 24945155 25830018 15632085 17994087 23185243 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 9288747 28648823 12364791 14747013 17210077 20075255 18362917 19820115 21719571 21347285 24508170 30249741 25581109 20798317 28992199 21282665 9215903 29403074 21993624 23258410 17975172 20809919 30723633 19393038 28087693
EMBL
KZ150142
PZC72932.1
RSAL01000051
RVE50207.1
AGBW02013840
OWR42679.1
+ More
NWSH01000499 PCG76006.1 KQ460473 KPJ14549.1 BABH01003117 BABH01003118 GANO01001209 JAB58662.1 GAMC01017243 JAB89312.1 CVRI01000014 CRK89686.1 GDHF01031607 GDHF01019297 JAI20707.1 JAI33017.1 CH477305 EAT44141.1 GAKP01014938 GAKP01014937 GAKP01014936 GAKP01014935 JAC44014.1 GAPW01001226 JAC12372.1 GFDL01000637 JAV34408.1 GBXI01013813 GBXI01011330 GBXI01008329 JAD00479.1 JAD02962.1 JAD05963.1 OUUW01000003 SPP77675.1 CH379064 EAL32226.1 KRT07068.1 CH902622 EDV34634.1 CH954180 EDV45832.1 CH933810 EDW06872.1 CP012528 ALC49409.1 CH964239 EDW82402.1 CM000366 EDX17092.1 AE014298 BT001403 BT010011 AAF45973.1 AAN09125.1 AAN71158.1 AAQ22480.1 CM000162 EDX01184.1 Y14162 CAA74575.1 CH940653 EDW62857.1 CH916376 EDV95353.1 UFQT01000095 SSX19702.1 AXCN02000277 NNAY01000126 OXU30723.1 AAAB01008987 EAA00976.6 NEVH01007404 PNF35785.1 KQ971312 EEZ98532.1 GEDC01009156 JAS28142.1 CH480819 EDW53144.1 KQ980530 KYN15604.1 GL888584 EGI59332.1 KQ976417 KYM89883.1 KQ981193 KYN45200.1 ADTU01027051 AJVK01008173 AJVK01008174 AJVK01008175 AJVK01008176 KQ978106 KYM96972.1 GFDF01003105 JAV10979.1 KK107324 EZA52529.1 KQ983203 KYQ46694.1 QOIP01000006 RLU22030.1 KC437299 AGC79112.1 GL445031 EFN60270.1 MF432991 ATL75342.1 KQ434946 KZC12357.1 KZ288387 PBC26417.1 LBMM01000512 KMQ98176.1 GL766312 EFZ15378.1 GU075990 ACY92519.1 AFYH01102853 AFYH01102854 AFYH01102855 AFYH01102856 GL446332 EFN88002.1 PYGN01000354 PSN47858.1 KQ414663 KOC65398.1 SAUD01001885 RXG70856.1 HAAF01002989 CCP74815.1 AEYP01077183 GBYB01006811 JAG76578.1 GEBF01006300 JAN97332.1 KN122676 KFO28858.1 AAKN02057765 AGTP01068828 KB030265 ELK19376.1 LWLT01000008 AANG04002859 AMGL01091635 MKHE01000009 OWK12104.1 AEMK02000012 AB192874 DQIR01207158 DQIR01279121 BAD89277.1 HDB62635.1 BC109826 AAI09827.1 GFFV01002354 JAV37591.1 GABZ01005728 JAA47797.1 AAQR03058869 NCKU01000394 RWS15661.1
NWSH01000499 PCG76006.1 KQ460473 KPJ14549.1 BABH01003117 BABH01003118 GANO01001209 JAB58662.1 GAMC01017243 JAB89312.1 CVRI01000014 CRK89686.1 GDHF01031607 GDHF01019297 JAI20707.1 JAI33017.1 CH477305 EAT44141.1 GAKP01014938 GAKP01014937 GAKP01014936 GAKP01014935 JAC44014.1 GAPW01001226 JAC12372.1 GFDL01000637 JAV34408.1 GBXI01013813 GBXI01011330 GBXI01008329 JAD00479.1 JAD02962.1 JAD05963.1 OUUW01000003 SPP77675.1 CH379064 EAL32226.1 KRT07068.1 CH902622 EDV34634.1 CH954180 EDV45832.1 CH933810 EDW06872.1 CP012528 ALC49409.1 CH964239 EDW82402.1 CM000366 EDX17092.1 AE014298 BT001403 BT010011 AAF45973.1 AAN09125.1 AAN71158.1 AAQ22480.1 CM000162 EDX01184.1 Y14162 CAA74575.1 CH940653 EDW62857.1 CH916376 EDV95353.1 UFQT01000095 SSX19702.1 AXCN02000277 NNAY01000126 OXU30723.1 AAAB01008987 EAA00976.6 NEVH01007404 PNF35785.1 KQ971312 EEZ98532.1 GEDC01009156 JAS28142.1 CH480819 EDW53144.1 KQ980530 KYN15604.1 GL888584 EGI59332.1 KQ976417 KYM89883.1 KQ981193 KYN45200.1 ADTU01027051 AJVK01008173 AJVK01008174 AJVK01008175 AJVK01008176 KQ978106 KYM96972.1 GFDF01003105 JAV10979.1 KK107324 EZA52529.1 KQ983203 KYQ46694.1 QOIP01000006 RLU22030.1 KC437299 AGC79112.1 GL445031 EFN60270.1 MF432991 ATL75342.1 KQ434946 KZC12357.1 KZ288387 PBC26417.1 LBMM01000512 KMQ98176.1 GL766312 EFZ15378.1 GU075990 ACY92519.1 AFYH01102853 AFYH01102854 AFYH01102855 AFYH01102856 GL446332 EFN88002.1 PYGN01000354 PSN47858.1 KQ414663 KOC65398.1 SAUD01001885 RXG70856.1 HAAF01002989 CCP74815.1 AEYP01077183 GBYB01006811 JAG76578.1 GEBF01006300 JAN97332.1 KN122676 KFO28858.1 AAKN02057765 AGTP01068828 KB030265 ELK19376.1 LWLT01000008 AANG04002859 AMGL01091635 MKHE01000009 OWK12104.1 AEMK02000012 AB192874 DQIR01207158 DQIR01279121 BAD89277.1 HDB62635.1 BC109826 AAI09827.1 GFFV01002354 JAV37591.1 GABZ01005728 JAA47797.1 AAQR03058869 NCKU01000394 RWS15661.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000053240
UP000005204
UP000095301
+ More
UP000183832 UP000008820 UP000268350 UP000001819 UP000007801 UP000008711 UP000095300 UP000192221 UP000009192 UP000092553 UP000007798 UP000000304 UP000000803 UP000002282 UP000008792 UP000001070 UP000075880 UP000075920 UP000075886 UP000215335 UP000007062 UP000235965 UP000002358 UP000007266 UP000001292 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000092462 UP000078542 UP000053097 UP000075809 UP000279307 UP000000311 UP000076502 UP000242457 UP000036403 UP000005203 UP000079169 UP000008672 UP000008237 UP000245037 UP000053825 UP000288706 UP000000715 UP000248482 UP000261640 UP000265300 UP000248483 UP000233160 UP000028990 UP000005447 UP000252040 UP000286640 UP000005215 UP000010552 UP000291000 UP000286641 UP000248484 UP000248481 UP000011712 UP000245341 UP000002356 UP000245340 UP000291022 UP000008227 UP000009136 UP000005225 UP000261681 UP000285301
UP000183832 UP000008820 UP000268350 UP000001819 UP000007801 UP000008711 UP000095300 UP000192221 UP000009192 UP000092553 UP000007798 UP000000304 UP000000803 UP000002282 UP000008792 UP000001070 UP000075880 UP000075920 UP000075886 UP000215335 UP000007062 UP000235965 UP000002358 UP000007266 UP000001292 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000092462 UP000078542 UP000053097 UP000075809 UP000279307 UP000000311 UP000076502 UP000242457 UP000036403 UP000005203 UP000079169 UP000008672 UP000008237 UP000245037 UP000053825 UP000288706 UP000000715 UP000248482 UP000261640 UP000265300 UP000248483 UP000233160 UP000028990 UP000005447 UP000252040 UP000286640 UP000005215 UP000010552 UP000291000 UP000286641 UP000248484 UP000248481 UP000011712 UP000245341 UP000002356 UP000245340 UP000291022 UP000008227 UP000009136 UP000005225 UP000261681 UP000285301
Interpro
IPR017986
WD40_repeat_dom
+ More
IPR024977 Apc4_WD40_dom
IPR001680 WD40_repeat
IPR033010 Cdc20/Fizzy
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR001932 PPM-type_phosphatase_dom
IPR000222 PP2C_BS
IPR036457 PPM-type_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR024977 Apc4_WD40_dom
IPR001680 WD40_repeat
IPR033010 Cdc20/Fizzy
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR001932 PPM-type_phosphatase_dom
IPR000222 PP2C_BS
IPR036457 PPM-type_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
Gene 3D
CDD
ProteinModelPortal
A0A2W1BCE8
A0A3S2NKX6
A0A212EMH9
A0A2A4JW45
A0A194RAM3
H9JEH7
+ More
U5EV22 A0A1I8MAB3 W8ASZ3 A0A1J1HT88 A0A0K8U1X0 Q17CQ5 A0A034VNG0 A0A023ESV6 A0A1Q3G3M5 A0A0A1X5L9 A0A3B0J7B4 Q29G84 B3MS79 B3NU56 A0A1I8NTT0 A0A1W4U5T1 B4L2J6 A0A0M3QZI0 B4NC43 B4R4P5 Q9W4H9 B4Q032 O18402 B4M7U0 B4JX77 A0A336LNN7 A0A182JCA7 A0A340TBF7 A0A182QYL6 A0A232FJF8 Q7PUU4 A0A2J7R4N4 K7ILS6 D6W9Q9 A0A1B6DR46 B4I0W3 A0A195DSV1 F4X2K7 A0A195BT79 A0A195FX20 A0A158NVA2 A0A1B0DPN4 A0A151IBE6 A0A1L8DX39 A0A026WBU6 A0A151WFR8 A0A3L8DNK8 M9PLJ4 E2B214 A0A291S6V5 A0A154PKT1 A0A2A3E3W2 A0A0J7L699 A0A087ZZZ4 E9IVM5 D1LX13 A0A3Q0IXN9 H3A259 E2B8G8 A0A2P8YUD0 A0A0L7R3J1 A0A444TLK5 U6CVQ8 M3XXP4 A0A0C9RIT7 A0A2Y9L6R5 A0A3Q3RQJ7 A0A340XFY9 A0A2Y9M2B6 A0A2K6FS10 A0A0P6J296 A0A091DEE4 A0A286XCI2 A0A341B2D0 A0A3Q7UP78 A0A287D3A8 L5L806 A0A452EMH9 A0A3Q7NB01 A0A2Y9TKN2 A0A2Y9I9T1 A0A337SP30 A0A2U3Y4Y4 W5PSG6 A0A2U3VKG9 A0A212D207 A0A452RRH7 Q5H7B9 Q32L05 A0A250Y1W1 K9ILH8 H0XAM8 A0A383ZNB5 A0A3S3SIC5
U5EV22 A0A1I8MAB3 W8ASZ3 A0A1J1HT88 A0A0K8U1X0 Q17CQ5 A0A034VNG0 A0A023ESV6 A0A1Q3G3M5 A0A0A1X5L9 A0A3B0J7B4 Q29G84 B3MS79 B3NU56 A0A1I8NTT0 A0A1W4U5T1 B4L2J6 A0A0M3QZI0 B4NC43 B4R4P5 Q9W4H9 B4Q032 O18402 B4M7U0 B4JX77 A0A336LNN7 A0A182JCA7 A0A340TBF7 A0A182QYL6 A0A232FJF8 Q7PUU4 A0A2J7R4N4 K7ILS6 D6W9Q9 A0A1B6DR46 B4I0W3 A0A195DSV1 F4X2K7 A0A195BT79 A0A195FX20 A0A158NVA2 A0A1B0DPN4 A0A151IBE6 A0A1L8DX39 A0A026WBU6 A0A151WFR8 A0A3L8DNK8 M9PLJ4 E2B214 A0A291S6V5 A0A154PKT1 A0A2A3E3W2 A0A0J7L699 A0A087ZZZ4 E9IVM5 D1LX13 A0A3Q0IXN9 H3A259 E2B8G8 A0A2P8YUD0 A0A0L7R3J1 A0A444TLK5 U6CVQ8 M3XXP4 A0A0C9RIT7 A0A2Y9L6R5 A0A3Q3RQJ7 A0A340XFY9 A0A2Y9M2B6 A0A2K6FS10 A0A0P6J296 A0A091DEE4 A0A286XCI2 A0A341B2D0 A0A3Q7UP78 A0A287D3A8 L5L806 A0A452EMH9 A0A3Q7NB01 A0A2Y9TKN2 A0A2Y9I9T1 A0A337SP30 A0A2U3Y4Y4 W5PSG6 A0A2U3VKG9 A0A212D207 A0A452RRH7 Q5H7B9 Q32L05 A0A250Y1W1 K9ILH8 H0XAM8 A0A383ZNB5 A0A3S3SIC5
PDB
5L9U
E-value=1.91276e-180,
Score=1625
Ontologies
GO
GO:0097027
GO:0010997
GO:1904668
GO:0010001
GO:0008347
GO:0005819
GO:0005680
GO:0061630
GO:0005813
GO:0005814
GO:0031536
GO:0030424
GO:0001745
GO:0031145
GO:0007455
GO:0030163
GO:0005737
GO:0007088
GO:0004721
GO:0046872
GO:0040020
GO:0045732
GO:0031965
GO:0090344
GO:0070306
GO:0072425
GO:0005654
GO:0008284
GO:1904145
GO:0051301
GO:0005515
GO:0008270
GO:0003676
GO:0007219
GO:0007275
GO:0050793
GO:0016876
GO:0043039
PANTHER
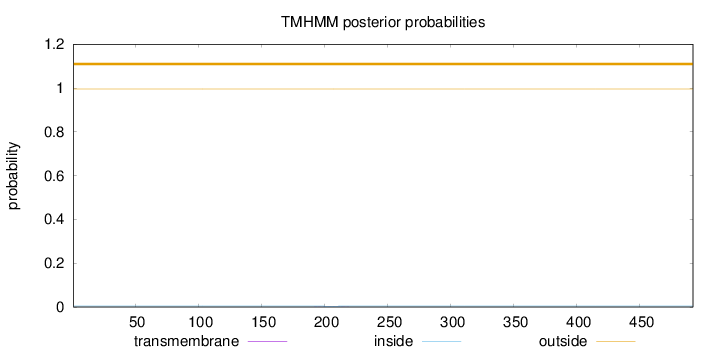
Topology
Length:
492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01578
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00432
outside
1 - 492
Population Genetic Test Statistics
Pi
252.63045
Theta
175.199766
Tajima's D
1.272913
CLR
0.079189
CSRT
0.735713214339283
Interpretation
Uncertain
