Gene
KWMTBOMO08692
Pre Gene Modal
BGIBMGA007926
Annotation
PREDICTED:_UDP-xylose_and_UDP-N-acetylglucosamine_transporter-like_isoform_X3_[Bombyx_mori]
Full name
UDP-xylose and UDP-N-acetylglucosamine transporter
Alternative Name
Solute carrier family 35 member B4
YEA4 homolog
YEA4 homolog
Location in the cell
PlasmaMembrane Reliability : 4.877
Sequence
CDS
ATGCCAGAGTGCGCTACTTTTATTACATTTCTTCAATTTTTATTCATAACTGTTCAAGGTTTTGTGGTAACAACAAAATTAGGAACAATTGCGCCAAAAATACCTTTAAAAAGCTATGGAATTTTGGTTACACTATTTTTCATATCAAGTGTAGCAAATAACTATGTCTATGCATTACATGTTCCATCCACTCTACATATGATTATAAGATCTGCTTCATCACCAGCCAGTTTGTTGGTCTATTGTTTAATAAAAAAACAAAAACCAAAACTGAACAAGACTGTAGGTTCGATTCTTATAAGTTTTGGGGTTGCTTTAGCTATGTATGGTGGTGCTCCCCCAACCGAGCAAAAAGGTAAATTCATTTATTGGTGCATAGGAGTCACTATATTGCTGTCAACACTGATAACCGGTGCATTCACTGGCCTTCAACAAGAAATACTTTACACAAAGTATGGGAAACATCCTGATGAGATGATGTTTTATACCCACGCAATACCTTTGCCCCTGTTTATTGGAATTTATCCCCAGCTGAAGAATATCTCCAAGAACATAAGTTTGGATGTCTGGGTCCTTATTATATTAAGTATAATTTCTCAATATTATTGTACACATTCTGTCCACAAGTTAGGTACAACAGAAACAACAGTGACTGTCACTTTTATATTAACCTTAAGAAAATTTGCATCCTTATTATTATCTTCTGTAGTTTTCACCAACAATCTCACTGTGTTTCATTGTATTGGGACTGTATTTGTAGCACTAGGCACTTATGTATATTTTGATTATTTTATTAGTAATAAACAAGTATCTGTAAGTCTTAAAGATAGATAG
Protein
MPECATFITFLQFLFITVQGFVVTTKLGTIAPKIPLKSYGILVTLFFISSVANNYVYALHVPSTLHMIIRSASSPASLLVYCLIKKQKPKLNKTVGSILISFGVALAMYGGAPPTEQKGKFIYWCIGVTILLSTLITGAFTGLQQEILYTKYGKHPDEMMFYTHAIPLPLFIGIYPQLKNISKNISLDVWVLIILSIISQYYCTHSVHKLGTTETTVTVTFILTLRKFASLLLSSVVFTNNLTVFHCIGTVFVALGTYVYFDYFISNKQVSVSLKDR
Summary
Description
Sugar transporter that specifically mediates the transport of UDP-xylose (UDP-Xyl) and UDP-N-acetylglucosamine (UDP-GlcNAc) from cytosol into Golgi.
Similarity
Belongs to the nucleotide-sugar transporter family. SLC35B subfamily.
Keywords
Complete proteome
Golgi apparatus
Membrane
Reference proteome
Sugar transport
Transmembrane
Transmembrane helix
Transport
Alternative splicing
Feature
chain UDP-xylose and UDP-N-acetylglucosamine transporter
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2A4JM92
A0A2H1W6G4
A0A1E1VXV8
A0A3S2PLV5
A0A194RF51
S4P631
+ More
D7EIK0 A0A0K8TQS5 J9KAW4 C4WUT8 W8CA71 A0A1U7R778 T1EDR4 A0A182FU17 A0A2M4ARB0 A0A2M4BT00 A7RT41 A0A2M4AQ20 A0A182U5H4 A0A2M3ZE64 T1DNH9 A0A182LPE1 A0A2M3Z9R0 A0A336K9S9 W5JJP8 A0A1Q3FHN4 A0A1W4X2Q0 A0A2C9GQT5 Q7Q357 A0A182XC69 R7UZV3 A0A2M4BT84 F1Q9W9 A0A182S6S7 V4B078 A0A182J086 A0A182LTW9 A0A1Q3FHT1 A0A1Q3FHF1 A0A1W4ZP54 A0A0L7QU21 A0A0N8K2T9 A0A084WB95 A0A069DSD8 D6CJI1 A0A2J8V3V0 Q5R8M3 Q7ZW46 A0A0L0CKV5 A0A3B4DGC8 B0WBU7 A0A182VGY5 Q174K4 W5UKZ0 G3QH86 A0A2R9BBZ5 H2R4Y0 Q969S0 H0V5U3 A0A2I3HMF3 A0A2K6SQ47 A0A2K5HAY1 A0A2K6DY13 A0A2K5ZU48 A0A0D9RAI6 A0A096NZN1 A0A2K5KME7 F7HQ89 Q95KB4 A0A2K5WEZ6 A0A2K6MC35 A0A1I8MZC4 A0A2R7X2L5 A0A3M0KDH6 U3D595 F1SNG8 A0A0T6BCG0 A0A2K6RVA9 A0A3Q1K3D6 A0A0K8T424 A0A1I8PEG5 A0A060VTM9 A0A2K5SJ99 T1P969 A0A1I8PE78 A0A2K5DGH0 M3V868 F7HWS3 B5X459 Q8CIA5 B5X3T7 E1BEH8 E2RAQ8 D3ZAN4 G3TAD3 A0A1D5NUD0 A0A452EYT5 A0A3Q7TAL0
D7EIK0 A0A0K8TQS5 J9KAW4 C4WUT8 W8CA71 A0A1U7R778 T1EDR4 A0A182FU17 A0A2M4ARB0 A0A2M4BT00 A7RT41 A0A2M4AQ20 A0A182U5H4 A0A2M3ZE64 T1DNH9 A0A182LPE1 A0A2M3Z9R0 A0A336K9S9 W5JJP8 A0A1Q3FHN4 A0A1W4X2Q0 A0A2C9GQT5 Q7Q357 A0A182XC69 R7UZV3 A0A2M4BT84 F1Q9W9 A0A182S6S7 V4B078 A0A182J086 A0A182LTW9 A0A1Q3FHT1 A0A1Q3FHF1 A0A1W4ZP54 A0A0L7QU21 A0A0N8K2T9 A0A084WB95 A0A069DSD8 D6CJI1 A0A2J8V3V0 Q5R8M3 Q7ZW46 A0A0L0CKV5 A0A3B4DGC8 B0WBU7 A0A182VGY5 Q174K4 W5UKZ0 G3QH86 A0A2R9BBZ5 H2R4Y0 Q969S0 H0V5U3 A0A2I3HMF3 A0A2K6SQ47 A0A2K5HAY1 A0A2K6DY13 A0A2K5ZU48 A0A0D9RAI6 A0A096NZN1 A0A2K5KME7 F7HQ89 Q95KB4 A0A2K5WEZ6 A0A2K6MC35 A0A1I8MZC4 A0A2R7X2L5 A0A3M0KDH6 U3D595 F1SNG8 A0A0T6BCG0 A0A2K6RVA9 A0A3Q1K3D6 A0A0K8T424 A0A1I8PEG5 A0A060VTM9 A0A2K5SJ99 T1P969 A0A1I8PE78 A0A2K5DGH0 M3V868 F7HWS3 B5X459 Q8CIA5 B5X3T7 E1BEH8 E2RAQ8 D3ZAN4 G3TAD3 A0A1D5NUD0 A0A452EYT5 A0A3Q7TAL0
Pubmed
26354079
23622113
18362917
19820115
26369729
24495485
+ More
23254933 17615350 20966253 20920257 23761445 12364791 14747013 17210077 23594743 24438588 26334808 21804562 26108605 17510324 23127152 22398555 22722832 16136131 15911612 14702039 16303743 12853948 12690205 15489334 21993624 17431167 25319552 25315136 25243066 25362486 24755649 20433749 16141072 19393038 16341006 15057822 15632090 15592404
23254933 17615350 20966253 20920257 23761445 12364791 14747013 17210077 23594743 24438588 26334808 21804562 26108605 17510324 23127152 22398555 22722832 16136131 15911612 14702039 16303743 12853948 12690205 15489334 21993624 17431167 25319552 25315136 25243066 25362486 24755649 20433749 16141072 19393038 16341006 15057822 15632090 15592404
EMBL
NWSH01001118
PCG72512.1
ODYU01006650
SOQ48681.1
GDQN01011488
JAT79566.1
+ More
RSAL01000001 RVE55175.1 KQ460473 KPJ14556.1 GAIX01007031 JAA85529.1 KQ971307 EFA12028.1 GDAI01000894 JAI16709.1 ABLF02031956 AK341218 BAH71658.1 GAMC01001541 GAMC01001540 GAMC01001539 JAC05017.1 AMQM01010107 KB097511 ESN95606.1 GGFK01009992 MBW43313.1 GGFJ01007075 MBW56216.1 DS469536 EDO45391.1 GGFK01009538 MBW42859.1 GGFM01006085 MBW26836.1 GAMD01002860 JAA98730.1 GGFM01004525 MBW25276.1 UFQS01000100 UFQT01000100 SSW99523.1 SSX19903.1 ADMH02001328 ETN62994.1 GFDL01007978 JAV27067.1 APCN01001930 AAAB01008966 EAA13009.4 AMQN01005494 KB296162 ELU12113.1 GGFJ01007073 MBW56214.1 CR855322 KB201037 ESO99426.1 AXCM01000360 GFDL01007919 JAV27126.1 GFDL01008112 JAV26933.1 KQ414740 KOC62054.1 JARO02000411 KPP78665.1 ATLV01022327 KE525331 KFB47489.1 GBGD01002243 JAC86646.1 FN825778 JH000366 CBL95112.1 EGV98807.1 NDHI03003435 PNJ52171.1 CR859728 BC045293 JRES01000340 KNC32109.1 DS231882 EDS42877.1 CH477409 EAT41500.1 EAT41501.1 JT418437 AHH42737.1 CABD030055224 AJFE02042763 AACZ04061431 AC187725 GABC01001180 GABC01001178 GABC01001177 GABF01005343 GABF01005342 GABF01005341 GABD01000381 GABD01000380 GABD01000379 GABD01000377 GABD01000376 GABD01000375 GABE01003699 GABE01003698 NBAG03000017 JAA10158.1 JAA16802.1 JAA32719.1 JAA41040.1 PNI99565.1 AJ971941 AB052892 AJ315497 AJ315498 AK027603 AK027726 AK075368 AK222874 AC008154 CH236950 CH471070 BC008413 AAKN02039167 ADFV01196483 ADFV01196484 ADFV01196485 AQIB01019288 AHZZ02023596 JSUE03029637 JU321386 JU321387 JU473667 JV048489 AFE65142.1 AFH30471.1 AFI38560.1 AB060201 AB063006 AQIA01048310 AQIA01048311 KK856567 PTY26032.1 QRBI01000112 RMC10551.1 RMC10558.1 GAMT01006932 GAMS01006924 GAMR01006660 GAMQ01006630 GAMP01009282 JAB04929.1 JAB16212.1 JAB27272.1 JAB35221.1 JAB43473.1 AEMK02000111 LJIG01002143 KRT84797.1 GBRD01005537 JAG60284.1 FR904298 CDQ58196.1 KA645249 AFP59878.1 GACC01000287 JAA74297.1 BT045828 ACI34090.1 AB041549 AK029969 AK033913 AK042150 AK138591 AK163299 BC033512 BT045706 ACI33968.1 AAEX03009230 AABR07060190 CH473959 EDM15298.1 AADN05000100 LWLT01000003
RSAL01000001 RVE55175.1 KQ460473 KPJ14556.1 GAIX01007031 JAA85529.1 KQ971307 EFA12028.1 GDAI01000894 JAI16709.1 ABLF02031956 AK341218 BAH71658.1 GAMC01001541 GAMC01001540 GAMC01001539 JAC05017.1 AMQM01010107 KB097511 ESN95606.1 GGFK01009992 MBW43313.1 GGFJ01007075 MBW56216.1 DS469536 EDO45391.1 GGFK01009538 MBW42859.1 GGFM01006085 MBW26836.1 GAMD01002860 JAA98730.1 GGFM01004525 MBW25276.1 UFQS01000100 UFQT01000100 SSW99523.1 SSX19903.1 ADMH02001328 ETN62994.1 GFDL01007978 JAV27067.1 APCN01001930 AAAB01008966 EAA13009.4 AMQN01005494 KB296162 ELU12113.1 GGFJ01007073 MBW56214.1 CR855322 KB201037 ESO99426.1 AXCM01000360 GFDL01007919 JAV27126.1 GFDL01008112 JAV26933.1 KQ414740 KOC62054.1 JARO02000411 KPP78665.1 ATLV01022327 KE525331 KFB47489.1 GBGD01002243 JAC86646.1 FN825778 JH000366 CBL95112.1 EGV98807.1 NDHI03003435 PNJ52171.1 CR859728 BC045293 JRES01000340 KNC32109.1 DS231882 EDS42877.1 CH477409 EAT41500.1 EAT41501.1 JT418437 AHH42737.1 CABD030055224 AJFE02042763 AACZ04061431 AC187725 GABC01001180 GABC01001178 GABC01001177 GABF01005343 GABF01005342 GABF01005341 GABD01000381 GABD01000380 GABD01000379 GABD01000377 GABD01000376 GABD01000375 GABE01003699 GABE01003698 NBAG03000017 JAA10158.1 JAA16802.1 JAA32719.1 JAA41040.1 PNI99565.1 AJ971941 AB052892 AJ315497 AJ315498 AK027603 AK027726 AK075368 AK222874 AC008154 CH236950 CH471070 BC008413 AAKN02039167 ADFV01196483 ADFV01196484 ADFV01196485 AQIB01019288 AHZZ02023596 JSUE03029637 JU321386 JU321387 JU473667 JV048489 AFE65142.1 AFH30471.1 AFI38560.1 AB060201 AB063006 AQIA01048310 AQIA01048311 KK856567 PTY26032.1 QRBI01000112 RMC10551.1 RMC10558.1 GAMT01006932 GAMS01006924 GAMR01006660 GAMQ01006630 GAMP01009282 JAB04929.1 JAB16212.1 JAB27272.1 JAB35221.1 JAB43473.1 AEMK02000111 LJIG01002143 KRT84797.1 GBRD01005537 JAG60284.1 FR904298 CDQ58196.1 KA645249 AFP59878.1 GACC01000287 JAA74297.1 BT045828 ACI34090.1 AB041549 AK029969 AK033913 AK042150 AK138591 AK163299 BC033512 BT045706 ACI33968.1 AAEX03009230 AABR07060190 CH473959 EDM15298.1 AADN05000100 LWLT01000003
Proteomes
UP000218220
UP000283053
UP000053240
UP000007266
UP000007819
UP000189706
+ More
UP000015101 UP000069272 UP000001593 UP000075902 UP000075882 UP000000673 UP000192223 UP000075840 UP000007062 UP000076407 UP000014760 UP000000437 UP000075901 UP000030746 UP000075880 UP000075883 UP000192224 UP000053825 UP000034805 UP000030765 UP000001075 UP000001595 UP000037069 UP000261440 UP000002320 UP000075903 UP000008820 UP000221080 UP000001519 UP000240080 UP000002277 UP000005640 UP000005447 UP000001073 UP000233220 UP000233080 UP000233120 UP000233140 UP000029965 UP000028761 UP000233060 UP000006718 UP000233100 UP000233180 UP000095301 UP000269221 UP000008227 UP000233200 UP000265040 UP000095300 UP000193380 UP000233040 UP000233020 UP000008225 UP000087266 UP000000589 UP000009136 UP000002254 UP000002494 UP000007646 UP000000539 UP000291000 UP000286640
UP000015101 UP000069272 UP000001593 UP000075902 UP000075882 UP000000673 UP000192223 UP000075840 UP000007062 UP000076407 UP000014760 UP000000437 UP000075901 UP000030746 UP000075880 UP000075883 UP000192224 UP000053825 UP000034805 UP000030765 UP000001075 UP000001595 UP000037069 UP000261440 UP000002320 UP000075903 UP000008820 UP000221080 UP000001519 UP000240080 UP000002277 UP000005640 UP000005447 UP000001073 UP000233220 UP000233080 UP000233120 UP000233140 UP000029965 UP000028761 UP000233060 UP000006718 UP000233100 UP000233180 UP000095301 UP000269221 UP000008227 UP000233200 UP000265040 UP000095300 UP000193380 UP000233040 UP000233020 UP000008225 UP000087266 UP000000589 UP000009136 UP000002254 UP000002494 UP000007646 UP000000539 UP000291000 UP000286640
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JM92
A0A2H1W6G4
A0A1E1VXV8
A0A3S2PLV5
A0A194RF51
S4P631
+ More
D7EIK0 A0A0K8TQS5 J9KAW4 C4WUT8 W8CA71 A0A1U7R778 T1EDR4 A0A182FU17 A0A2M4ARB0 A0A2M4BT00 A7RT41 A0A2M4AQ20 A0A182U5H4 A0A2M3ZE64 T1DNH9 A0A182LPE1 A0A2M3Z9R0 A0A336K9S9 W5JJP8 A0A1Q3FHN4 A0A1W4X2Q0 A0A2C9GQT5 Q7Q357 A0A182XC69 R7UZV3 A0A2M4BT84 F1Q9W9 A0A182S6S7 V4B078 A0A182J086 A0A182LTW9 A0A1Q3FHT1 A0A1Q3FHF1 A0A1W4ZP54 A0A0L7QU21 A0A0N8K2T9 A0A084WB95 A0A069DSD8 D6CJI1 A0A2J8V3V0 Q5R8M3 Q7ZW46 A0A0L0CKV5 A0A3B4DGC8 B0WBU7 A0A182VGY5 Q174K4 W5UKZ0 G3QH86 A0A2R9BBZ5 H2R4Y0 Q969S0 H0V5U3 A0A2I3HMF3 A0A2K6SQ47 A0A2K5HAY1 A0A2K6DY13 A0A2K5ZU48 A0A0D9RAI6 A0A096NZN1 A0A2K5KME7 F7HQ89 Q95KB4 A0A2K5WEZ6 A0A2K6MC35 A0A1I8MZC4 A0A2R7X2L5 A0A3M0KDH6 U3D595 F1SNG8 A0A0T6BCG0 A0A2K6RVA9 A0A3Q1K3D6 A0A0K8T424 A0A1I8PEG5 A0A060VTM9 A0A2K5SJ99 T1P969 A0A1I8PE78 A0A2K5DGH0 M3V868 F7HWS3 B5X459 Q8CIA5 B5X3T7 E1BEH8 E2RAQ8 D3ZAN4 G3TAD3 A0A1D5NUD0 A0A452EYT5 A0A3Q7TAL0
D7EIK0 A0A0K8TQS5 J9KAW4 C4WUT8 W8CA71 A0A1U7R778 T1EDR4 A0A182FU17 A0A2M4ARB0 A0A2M4BT00 A7RT41 A0A2M4AQ20 A0A182U5H4 A0A2M3ZE64 T1DNH9 A0A182LPE1 A0A2M3Z9R0 A0A336K9S9 W5JJP8 A0A1Q3FHN4 A0A1W4X2Q0 A0A2C9GQT5 Q7Q357 A0A182XC69 R7UZV3 A0A2M4BT84 F1Q9W9 A0A182S6S7 V4B078 A0A182J086 A0A182LTW9 A0A1Q3FHT1 A0A1Q3FHF1 A0A1W4ZP54 A0A0L7QU21 A0A0N8K2T9 A0A084WB95 A0A069DSD8 D6CJI1 A0A2J8V3V0 Q5R8M3 Q7ZW46 A0A0L0CKV5 A0A3B4DGC8 B0WBU7 A0A182VGY5 Q174K4 W5UKZ0 G3QH86 A0A2R9BBZ5 H2R4Y0 Q969S0 H0V5U3 A0A2I3HMF3 A0A2K6SQ47 A0A2K5HAY1 A0A2K6DY13 A0A2K5ZU48 A0A0D9RAI6 A0A096NZN1 A0A2K5KME7 F7HQ89 Q95KB4 A0A2K5WEZ6 A0A2K6MC35 A0A1I8MZC4 A0A2R7X2L5 A0A3M0KDH6 U3D595 F1SNG8 A0A0T6BCG0 A0A2K6RVA9 A0A3Q1K3D6 A0A0K8T424 A0A1I8PEG5 A0A060VTM9 A0A2K5SJ99 T1P969 A0A1I8PE78 A0A2K5DGH0 M3V868 F7HWS3 B5X459 Q8CIA5 B5X3T7 E1BEH8 E2RAQ8 D3ZAN4 G3TAD3 A0A1D5NUD0 A0A452EYT5 A0A3Q7TAL0
Ontologies
GO
PANTHER
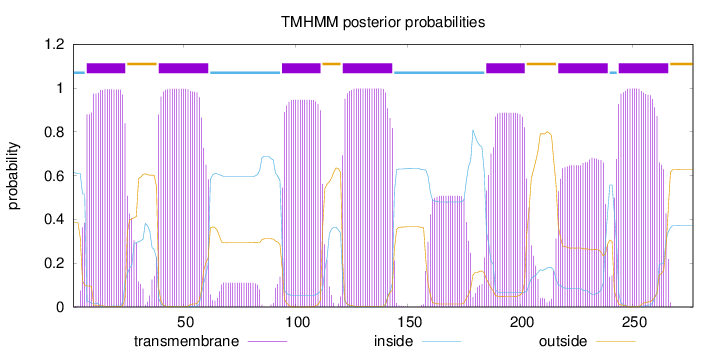
Topology
Subcellular location
Golgi apparatus membrane
Length:
277
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
148.41902
Exp number, first 60 AAs:
41.8662
Total prob of N-in:
0.61532
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 24
outside
25 - 38
TMhelix
39 - 61
inside
62 - 93
TMhelix
94 - 111
outside
112 - 120
TMhelix
121 - 143
inside
144 - 184
TMhelix
185 - 202
outside
203 - 216
TMhelix
217 - 239
inside
240 - 243
TMhelix
244 - 266
outside
267 - 277
Population Genetic Test Statistics
Pi
243.188369
Theta
211.372985
Tajima's D
0.435756
CLR
0.513763
CSRT
0.492575371231438
Interpretation
Uncertain
