Gene
KWMTBOMO08690
Pre Gene Modal
BGIBMGA007927
Annotation
PREDICTED:_neuropathy_target_esterase_sws_isoform_X2_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.66
Sequence
CDS
ATGGATGTAGTGGATCTATTGAGCAACATAAACGATAAAGCGAACATGTTCGCTGTCACGACTTGGACATCAGAATGGTCGAATAATTTCCAAGAGAATCAATTGCTGTGGTCTTTCTGTGGCTGCCTTTTAGTATCCTTGCTGATCGTATTCTTTTATTACTATAGGAGATGGCGAGTCAAAGAATTAGTTGGCGATGTCTGCGCGTCAGGGGCTGCAGGGGAGCCGGCGAAGCGTTTCAGAAAACGCGACAAGATGATGTTCTACGGTAGACGTATGTTAAGAAAAGTCAAGTCCATATCAAACTCTGGTCAGGGTCGCAAGAGGAGAGCTGTCATGAGGTTCGCGCGTAAACTCCTACAGCTCAAAAAAGACTCCGCTCCTGAACAACTTAAGGTACTGGAACCACCAGCAGAATACTTGGAAGAGGACCTCACAAGTGACGATCGAGTTCCACCGGATGCTCTATACATGCTGCACAGCATCAGAGTTTTCGGGCACTTCGAGAAACCAGTCTTCCTAATGCTTTGTAAACACACTGAGATACTGAACTTGCCTGCTGGCTCATTCCTGTTCAAAGTTGGTGACACCGACGAAAATGTATACGTGGTTCAAACTGGTCGCGTGAACGTATACATCACGAATCAAGATGGAAGCAGTCTCTCGTTGAAGGTCGTCAGGGCGGGAGAGAGCGTCACGTCTTTGCTTAGCTTTACTGATGTTCTCACGGGTCACTCGCAACCTTACAAGACAGTGAACGCCAAAGCTCTTGAAGATTCGCAAGTCATAAAGCTGCCAATGAGAGCTTTCCAAGAAGTATTCAAAGAGTACCCCGACATATTCGTCAGGGTAATACAGATCATCATGGTTCGTCTTCAGAGGGTAACGTTCACAGCTCTTCACCAGTACCTTGGCTTGAGTGCTGAACTTGTAAATCCTGGCAAAGAGAAACGTCGACCGATGACGGTTTCGTCGTCTCCAGGGAAGACAGCAAAGGCGCCAGTTGACAACCTGTCGCACTCCCCGCACCCCGCCGACAAGTCCACCCCGGATCACGTGGAGGGAGGGCATCAAGTCTCCTCGCCCATCCACATCCAGGGTCGGCGTCAGAGGCCCGACATGGTGCCCGACGTAACATCGAACACACTGCAGCAGCCGGACGTGCAACCGACATCCTCGCTGTCGCGCAGCAAGGAGGGCAGTTCCTCGTTCAAGAAGCACATCAATGACAACCTAGACGAGAAGGCATTAACTCAAATAGCAACCGACGCGTTCGTGAAGGAGCTGGGTCTGGAGAACGAGCAGCTCTTGCAGGGGAGCGTCCAAGTGCGGGACCTGCCCGCCGGGACCTACATCATGAAGGAGGAGAGCCATAAGGACGTGGCATTGGTATATTTGCTATCCGGAGCTCTGCTGGTGTCACAGAGAGTGGCTGAAGGTGAGGGCGAAGTGCACATGTTCACAGCCTATCCAGGTGAAGTGGAAGGCGGACTTGCAGTTCTAACGGGCGAGCCAAGTTTCTTTTCTATACGAGCGAAACACTTCTCGAGGATCGGTCTGCTGTCCAAGACCACTGTGTACAGCATCATGAGAGAAAGACCCTCGGTGGTGTTGCACATCGCCAACACCGTCGTGCGGAGACTGTCGCCGTTCGTGCGCCAAGTGGACTTCGCTCTGGATTGGGTGAGACCCTTAAAACAAATATTAGATCAAACACACCGGCTTTTCTTCCAGGAACAACTATTAATGTTGATATAG
Protein
MDVVDLLSNINDKANMFAVTTWTSEWSNNFQENQLLWSFCGCLLVSLLIVFFYYYRRWRVKELVGDVCASGAAGEPAKRFRKRDKMMFYGRRMLRKVKSISNSGQGRKRRAVMRFARKLLQLKKDSAPEQLKVLEPPAEYLEEDLTSDDRVPPDALYMLHSIRVFGHFEKPVFLMLCKHTEILNLPAGSFLFKVGDTDENVYVVQTGRVNVYITNQDGSSLSLKVVRAGESVTSLLSFTDVLTGHSQPYKTVNAKALEDSQVIKLPMRAFQEVFKEYPDIFVRVIQIIMVRLQRVTFTALHQYLGLSAELVNPGKEKRRPMTVSSSPGKTAKAPVDNLSHSPHPADKSTPDHVEGGHQVSSPIHIQGRRQRPDMVPDVTSNTLQQPDVQPTSSLSRSKEGSSSFKKHINDNLDEKALTQIATDAFVKELGLENEQLLQGSVQVRDLPAGTYIMKEESHKDVALVYLLSGALLVSQRVAEGEGEVHMFTAYPGEVEGGLAVLTGEPSFFSIRAKHFSRIGLLSKTTVYSIMRERPSVVLHIANTVVRRLSPFVRQVDFALDWVRPLKQILDQTHRLFFQEQLLMLI
Summary
Uniprot
A0A2A4JKD9
A0A2W1BNY1
A0A194PPP5
H9JEI0
A0A2A4JL68
A0A3S2MAJ0
+ More
A0A2H1W600 A0A2A4JKB4 A0A2A4JLR8 A0A2A4JM99 A0A194RAN7 A0A2J7QV89 A0A158NV62 A0A195BQH0 A0A2J7QV91 A0A2J7QV86 A0A0C9RHG8 A0A195F595 K7IQ10 A0A151XE85 A0A151JN95 F4W4X6 V9IBX3 A0A232F867 A0A088ACE8 A0A026WPN4 A0A067RS17 A0A2A3EFX0 D6W9Q7 A0A1B6MDX6 A0A139WN47 A0A0M9A8F9 A0A1B6F290 V5GPU5 A0A023F3T3 A0A0V0G706 A0A1B6E580 A0A1B6CYY3 A0A1B6HT09 A0A3L8DN19 T1HIF7 A0A0T6AX92 A0A0J7L6P9 A0A195CH99 A0A1Y1MM72 A0A0L7R5B4 A0A1Y1MJA9 A0A0A9XIY4 A0A1Y1MJ21 A0A1Y1MMU8 A0A0A9XBN8 A0A1Y1MPB8 E2A0X2 A0A1Y1MJ22 A0A154PJP7 U4UGD9 A0A310SL74 A0A182J5J9 J9K997 A0A1Q3FE81 N6U063 E2BKL4 B0W7V2 A0A182GNA6 A0A182W5J3 A0A084VLT2 A0A182XDV8 A0A182RHI3 A0A3F2Z1K2 A0A182IAP6 A0A1S4G951 A0A182XXS1 A0A3F2YXS0 A0A1B0FAA6 A0A336MD57 A0A1J1IYU6 A0A1J1IVY4 A0A182QB96 A0A182MPR2 A0A1B0CSP7 A0A0P5FLP0 A0A0P5CKW4 A0A0P5DAW4 A0A0P6H263 A0A0N8ADV2 A0A0P5M2U8 A0A0P5ZW03 A0A1B0D0J3 A0A0P5DV65 A0A0P6GFW6 A0A0P5NYB1 A0A0N8B395 A0A0P5DD26 A0A0P5NRI5 A0A0N8DG32 A0A0P6A5D2 A0A0P5MSZ2 A0A0N8ABI4 A0A0P6GG88 A0A0P6AQM4
A0A2H1W600 A0A2A4JKB4 A0A2A4JLR8 A0A2A4JM99 A0A194RAN7 A0A2J7QV89 A0A158NV62 A0A195BQH0 A0A2J7QV91 A0A2J7QV86 A0A0C9RHG8 A0A195F595 K7IQ10 A0A151XE85 A0A151JN95 F4W4X6 V9IBX3 A0A232F867 A0A088ACE8 A0A026WPN4 A0A067RS17 A0A2A3EFX0 D6W9Q7 A0A1B6MDX6 A0A139WN47 A0A0M9A8F9 A0A1B6F290 V5GPU5 A0A023F3T3 A0A0V0G706 A0A1B6E580 A0A1B6CYY3 A0A1B6HT09 A0A3L8DN19 T1HIF7 A0A0T6AX92 A0A0J7L6P9 A0A195CH99 A0A1Y1MM72 A0A0L7R5B4 A0A1Y1MJA9 A0A0A9XIY4 A0A1Y1MJ21 A0A1Y1MMU8 A0A0A9XBN8 A0A1Y1MPB8 E2A0X2 A0A1Y1MJ22 A0A154PJP7 U4UGD9 A0A310SL74 A0A182J5J9 J9K997 A0A1Q3FE81 N6U063 E2BKL4 B0W7V2 A0A182GNA6 A0A182W5J3 A0A084VLT2 A0A182XDV8 A0A182RHI3 A0A3F2Z1K2 A0A182IAP6 A0A1S4G951 A0A182XXS1 A0A3F2YXS0 A0A1B0FAA6 A0A336MD57 A0A1J1IYU6 A0A1J1IVY4 A0A182QB96 A0A182MPR2 A0A1B0CSP7 A0A0P5FLP0 A0A0P5CKW4 A0A0P5DAW4 A0A0P6H263 A0A0N8ADV2 A0A0P5M2U8 A0A0P5ZW03 A0A1B0D0J3 A0A0P5DV65 A0A0P6GFW6 A0A0P5NYB1 A0A0N8B395 A0A0P5DD26 A0A0P5NRI5 A0A0N8DG32 A0A0P6A5D2 A0A0P5MSZ2 A0A0N8ABI4 A0A0P6GG88 A0A0P6AQM4
Pubmed
EMBL
NWSH01001118
PCG72517.1
KZ150050
PZC74430.1
KQ459596
KPI95416.1
+ More
BABH01003154 BABH01003155 PCG72516.1 RSAL01000001 RVE55177.1 ODYU01006561 SOQ48510.1 PCG72515.1 PCG72514.1 PCG72520.1 KQ460473 KPJ14559.1 NEVH01010478 PNF32499.1 ADTU01026908 ADTU01026909 ADTU01026910 KQ976424 KYM88209.1 PNF32497.1 PNF32498.1 GBYB01006371 JAG76138.1 KQ981820 KYN35244.1 KQ982254 KYQ58682.1 KQ978850 KYN27955.1 GL887596 EGI70672.1 JR039172 JR039173 AEY58564.1 AEY58565.1 NNAY01000723 OXU26792.1 KK107135 EZA58017.1 KK852460 KDR23520.1 KZ288256 PBC30620.1 KQ971312 EEZ98531.2 GEBQ01005873 JAT34104.1 KYB29225.1 KQ435712 KOX79375.1 GECZ01025400 JAS44369.1 GALX01002317 JAB66149.1 GBBI01002661 JAC16051.1 GECL01002914 JAP03210.1 GEDC01004208 JAS33090.1 GEDC01018845 GEDC01013659 JAS18453.1 JAS23639.1 GECU01029900 JAS77806.1 QOIP01000006 RLU21676.1 ACPB03001249 LJIG01022626 KRT79563.1 LBMM01000443 KMQ98341.1 KQ977827 KYM99458.1 GEZM01030137 GEZM01030134 JAV85790.1 KQ414654 KOC66038.1 GEZM01030136 JAV85791.1 GBHO01026549 GBRD01007580 GDHC01009539 JAG17055.1 JAG58241.1 JAQ09090.1 GEZM01030131 GEZM01030130 JAV85799.1 GEZM01030133 JAV85795.1 GBHO01026548 GBRD01007579 JAG17056.1 JAG58242.1 GEZM01030138 GEZM01030132 JAV85796.1 GL435626 EFN73025.1 GEZM01030135 GEZM01030129 JAV85792.1 KQ434923 KZC11508.1 KB632309 ERL92097.1 KQ760132 OAD61817.1 ABLF02030155 GFDL01009172 JAV25873.1 APGK01044466 KB741026 ENN74930.1 GL448819 EFN83712.1 DS231855 EDS38246.1 JXUM01076204 JXUM01076205 JXUM01076206 KQ562931 KXJ74816.1 ATLV01014551 KE524974 KFB38926.1 APCN01001296 AAAB01008846 CCAG010020857 CCAG010020858 UFQT01000668 SSX26327.1 CVRI01000063 CRL04334.1 CRL04335.1 AXCN02002156 AXCM01003763 AJWK01026359 GDIQ01253572 JAJ98152.1 GDIP01169013 JAJ54389.1 GDIP01162719 JAJ60683.1 GDIQ01035898 JAN58839.1 GDIP01157121 JAJ66281.1 GDIQ01170297 JAK81428.1 GDIP01038589 JAM65126.1 AJVK01021458 AJVK01021459 GDIP01157120 JAJ66282.1 GDIQ01033698 JAN61039.1 GDIQ01157456 JAK94269.1 GDIQ01217099 JAK34626.1 GDIP01161768 JAJ61634.1 GDIP01251908 GDIQ01145420 JAI71493.1 JAL06306.1 GDIP01037191 JAM66524.1 GDIP01034690 JAM69025.1 GDIQ01174389 JAK77336.1 GDIP01163665 JAJ59737.1 GDIQ01033697 JAN61040.1 GDIP01039634 JAM64081.1
BABH01003154 BABH01003155 PCG72516.1 RSAL01000001 RVE55177.1 ODYU01006561 SOQ48510.1 PCG72515.1 PCG72514.1 PCG72520.1 KQ460473 KPJ14559.1 NEVH01010478 PNF32499.1 ADTU01026908 ADTU01026909 ADTU01026910 KQ976424 KYM88209.1 PNF32497.1 PNF32498.1 GBYB01006371 JAG76138.1 KQ981820 KYN35244.1 KQ982254 KYQ58682.1 KQ978850 KYN27955.1 GL887596 EGI70672.1 JR039172 JR039173 AEY58564.1 AEY58565.1 NNAY01000723 OXU26792.1 KK107135 EZA58017.1 KK852460 KDR23520.1 KZ288256 PBC30620.1 KQ971312 EEZ98531.2 GEBQ01005873 JAT34104.1 KYB29225.1 KQ435712 KOX79375.1 GECZ01025400 JAS44369.1 GALX01002317 JAB66149.1 GBBI01002661 JAC16051.1 GECL01002914 JAP03210.1 GEDC01004208 JAS33090.1 GEDC01018845 GEDC01013659 JAS18453.1 JAS23639.1 GECU01029900 JAS77806.1 QOIP01000006 RLU21676.1 ACPB03001249 LJIG01022626 KRT79563.1 LBMM01000443 KMQ98341.1 KQ977827 KYM99458.1 GEZM01030137 GEZM01030134 JAV85790.1 KQ414654 KOC66038.1 GEZM01030136 JAV85791.1 GBHO01026549 GBRD01007580 GDHC01009539 JAG17055.1 JAG58241.1 JAQ09090.1 GEZM01030131 GEZM01030130 JAV85799.1 GEZM01030133 JAV85795.1 GBHO01026548 GBRD01007579 JAG17056.1 JAG58242.1 GEZM01030138 GEZM01030132 JAV85796.1 GL435626 EFN73025.1 GEZM01030135 GEZM01030129 JAV85792.1 KQ434923 KZC11508.1 KB632309 ERL92097.1 KQ760132 OAD61817.1 ABLF02030155 GFDL01009172 JAV25873.1 APGK01044466 KB741026 ENN74930.1 GL448819 EFN83712.1 DS231855 EDS38246.1 JXUM01076204 JXUM01076205 JXUM01076206 KQ562931 KXJ74816.1 ATLV01014551 KE524974 KFB38926.1 APCN01001296 AAAB01008846 CCAG010020857 CCAG010020858 UFQT01000668 SSX26327.1 CVRI01000063 CRL04334.1 CRL04335.1 AXCN02002156 AXCM01003763 AJWK01026359 GDIQ01253572 JAJ98152.1 GDIP01169013 JAJ54389.1 GDIP01162719 JAJ60683.1 GDIQ01035898 JAN58839.1 GDIP01157121 JAJ66281.1 GDIQ01170297 JAK81428.1 GDIP01038589 JAM65126.1 AJVK01021458 AJVK01021459 GDIP01157120 JAJ66282.1 GDIQ01033698 JAN61039.1 GDIQ01157456 JAK94269.1 GDIQ01217099 JAK34626.1 GDIP01161768 JAJ61634.1 GDIP01251908 GDIQ01145420 JAI71493.1 JAL06306.1 GDIP01037191 JAM66524.1 GDIP01034690 JAM69025.1 GDIQ01174389 JAK77336.1 GDIP01163665 JAJ59737.1 GDIQ01033697 JAN61040.1 GDIP01039634 JAM64081.1
Proteomes
UP000218220
UP000053268
UP000005204
UP000283053
UP000053240
UP000235965
+ More
UP000005205 UP000078540 UP000078541 UP000002358 UP000075809 UP000078492 UP000007755 UP000215335 UP000005203 UP000053097 UP000027135 UP000242457 UP000007266 UP000053105 UP000279307 UP000015103 UP000036403 UP000078542 UP000053825 UP000000311 UP000076502 UP000030742 UP000075880 UP000007819 UP000019118 UP000008237 UP000002320 UP000069940 UP000249989 UP000075920 UP000030765 UP000076407 UP000075900 UP000075840 UP000076408 UP000092444 UP000183832 UP000075886 UP000075883 UP000092461 UP000092462
UP000005205 UP000078540 UP000078541 UP000002358 UP000075809 UP000078492 UP000007755 UP000215335 UP000005203 UP000053097 UP000027135 UP000242457 UP000007266 UP000053105 UP000279307 UP000015103 UP000036403 UP000078542 UP000053825 UP000000311 UP000076502 UP000030742 UP000075880 UP000007819 UP000019118 UP000008237 UP000002320 UP000069940 UP000249989 UP000075920 UP000030765 UP000076407 UP000075900 UP000075840 UP000076408 UP000092444 UP000183832 UP000075886 UP000075883 UP000092461 UP000092462
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JKD9
A0A2W1BNY1
A0A194PPP5
H9JEI0
A0A2A4JL68
A0A3S2MAJ0
+ More
A0A2H1W600 A0A2A4JKB4 A0A2A4JLR8 A0A2A4JM99 A0A194RAN7 A0A2J7QV89 A0A158NV62 A0A195BQH0 A0A2J7QV91 A0A2J7QV86 A0A0C9RHG8 A0A195F595 K7IQ10 A0A151XE85 A0A151JN95 F4W4X6 V9IBX3 A0A232F867 A0A088ACE8 A0A026WPN4 A0A067RS17 A0A2A3EFX0 D6W9Q7 A0A1B6MDX6 A0A139WN47 A0A0M9A8F9 A0A1B6F290 V5GPU5 A0A023F3T3 A0A0V0G706 A0A1B6E580 A0A1B6CYY3 A0A1B6HT09 A0A3L8DN19 T1HIF7 A0A0T6AX92 A0A0J7L6P9 A0A195CH99 A0A1Y1MM72 A0A0L7R5B4 A0A1Y1MJA9 A0A0A9XIY4 A0A1Y1MJ21 A0A1Y1MMU8 A0A0A9XBN8 A0A1Y1MPB8 E2A0X2 A0A1Y1MJ22 A0A154PJP7 U4UGD9 A0A310SL74 A0A182J5J9 J9K997 A0A1Q3FE81 N6U063 E2BKL4 B0W7V2 A0A182GNA6 A0A182W5J3 A0A084VLT2 A0A182XDV8 A0A182RHI3 A0A3F2Z1K2 A0A182IAP6 A0A1S4G951 A0A182XXS1 A0A3F2YXS0 A0A1B0FAA6 A0A336MD57 A0A1J1IYU6 A0A1J1IVY4 A0A182QB96 A0A182MPR2 A0A1B0CSP7 A0A0P5FLP0 A0A0P5CKW4 A0A0P5DAW4 A0A0P6H263 A0A0N8ADV2 A0A0P5M2U8 A0A0P5ZW03 A0A1B0D0J3 A0A0P5DV65 A0A0P6GFW6 A0A0P5NYB1 A0A0N8B395 A0A0P5DD26 A0A0P5NRI5 A0A0N8DG32 A0A0P6A5D2 A0A0P5MSZ2 A0A0N8ABI4 A0A0P6GG88 A0A0P6AQM4
A0A2H1W600 A0A2A4JKB4 A0A2A4JLR8 A0A2A4JM99 A0A194RAN7 A0A2J7QV89 A0A158NV62 A0A195BQH0 A0A2J7QV91 A0A2J7QV86 A0A0C9RHG8 A0A195F595 K7IQ10 A0A151XE85 A0A151JN95 F4W4X6 V9IBX3 A0A232F867 A0A088ACE8 A0A026WPN4 A0A067RS17 A0A2A3EFX0 D6W9Q7 A0A1B6MDX6 A0A139WN47 A0A0M9A8F9 A0A1B6F290 V5GPU5 A0A023F3T3 A0A0V0G706 A0A1B6E580 A0A1B6CYY3 A0A1B6HT09 A0A3L8DN19 T1HIF7 A0A0T6AX92 A0A0J7L6P9 A0A195CH99 A0A1Y1MM72 A0A0L7R5B4 A0A1Y1MJA9 A0A0A9XIY4 A0A1Y1MJ21 A0A1Y1MMU8 A0A0A9XBN8 A0A1Y1MPB8 E2A0X2 A0A1Y1MJ22 A0A154PJP7 U4UGD9 A0A310SL74 A0A182J5J9 J9K997 A0A1Q3FE81 N6U063 E2BKL4 B0W7V2 A0A182GNA6 A0A182W5J3 A0A084VLT2 A0A182XDV8 A0A182RHI3 A0A3F2Z1K2 A0A182IAP6 A0A1S4G951 A0A182XXS1 A0A3F2YXS0 A0A1B0FAA6 A0A336MD57 A0A1J1IYU6 A0A1J1IVY4 A0A182QB96 A0A182MPR2 A0A1B0CSP7 A0A0P5FLP0 A0A0P5CKW4 A0A0P5DAW4 A0A0P6H263 A0A0N8ADV2 A0A0P5M2U8 A0A0P5ZW03 A0A1B0D0J3 A0A0P5DV65 A0A0P6GFW6 A0A0P5NYB1 A0A0N8B395 A0A0P5DD26 A0A0P5NRI5 A0A0N8DG32 A0A0P6A5D2 A0A0P5MSZ2 A0A0N8ABI4 A0A0P6GG88 A0A0P6AQM4
PDB
6DT4
E-value=0.00298018,
Score=98
Ontologies
KEGG
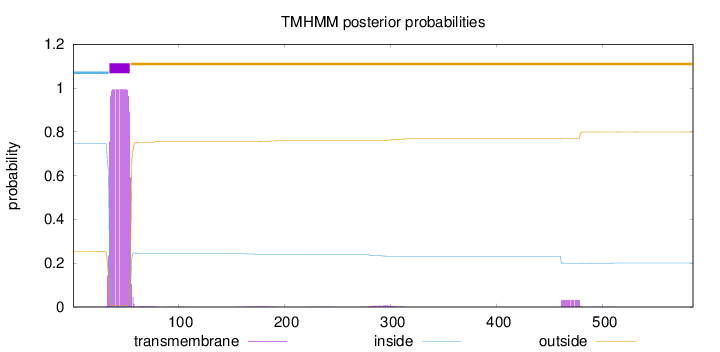
Topology
Length:
585
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.51544
Exp number, first 60 AAs:
20.61549
Total prob of N-in:
0.74640
POSSIBLE N-term signal
sequence
inside
1 - 34
TMhelix
35 - 54
outside
55 - 585
Population Genetic Test Statistics
Pi
236.620621
Theta
177.326754
Tajima's D
1.210562
CLR
0.156393
CSRT
0.714364281785911
Interpretation
Uncertain
