Pre Gene Modal
BGIBMGA007928
Annotation
PREDICTED:_neuropathy_target_esterase_sws_isoform_X3_[Bombyx_mori]
Full name
Neuropathy target esterase sws
Alternative Name
Swiss cheese
Location in the cell
Mitochondrial Reliability : 1.467
Sequence
CDS
ATGGTGACGCAGACTCGTCGGAGCACAACGGTGATGGCGGTGCGGGACTCGGAACTGGCGAAGCTGCCCGAAGGCCTCTTCAACGCCATCAAGCTGAGGTTCCCGGTCGTCGTGACTAGGCTGATCAACTTGCTGGGACATAGGATTCTAGGTTCCTGGCAGAAGCCGACGCGGAGTCTGGGCGGCGCGCCCGCGATCGACTCGAGGCCGTTGCAGCACAACTTCAGCACGGTGGCCGTGGTGCCCGTCAGCGACGACGTGCCCCTCACCGCCTTCACCTACGAGCTGTACCACTCGCTCTGCGCCATAGGTCCAACGGTCCGTCTCACTTCGGATGTCATAAGGAAACTGCTCGGTCTAACGATAATGGACCCGAATAACGAGTACAGACTAAGCTCTTGGCTGGCGCAACAAGAAGATAAACACAAGGTGGCGCTGTACCAGTGCGACCCGAGCCTCACGCAGTGGACGCAGCGCTGTATCCGGCAGGCCGACTGCATACTGATCGTGGCGCTCGGCGACAAGCAGCCCAGCATCGGGAAGATCGAGAAGGAAATCGAACGTCTAGCCATACGAACTCAGAAGGAGCTAGTTCTTCTGCACCGGGAGGGCGGACCGAACCCCTCCGGCACCGTGCACTGGCTCAACATGCGCACCTGGGTCAGCCAGCACCACCACGTGAGGTGTCCGCATCGAATGTTCACAAGGAAAAGCCAGTACCGGATTAGCGAGCTGTACAGCAAGGTGCTGATGTCTGAGGCCAACGTGCACTCGGACTTCAGTCGGCTGGCGCGCTGGCTCACCGCCACCGCCGTGGGGCTGGTGCTGGGGGGCGGAGGCGCGCGGGGGGCGGCGCACGTCGGCATGATCCGCGCCATACAGGAAGCAGGAATCCCAATAGATATGGTGGGGGGCGTCAGCATCGGCGCGTTTATGGGCGCCTTGTGGTGCATGGAGAGGAACATCACCACTGTCACGCAGAAGGCCAGGGAATGGTCCAAGAAAATGACGCAATGGGGTAGGCAGCTTCTAGACTTGACGTACCCAGCCACGTCGATGTTCTCCGGGCGACAGTTCAATGCCACCATCAGGGCGACGTTTGGCGAGGTTCACATCGAGGACCTGTGGCTGCCATACTTCACTGTCACCACTGACATTAGTTCCAGTTGCATGAGAATCCACAGGCACGGTTCGCTGTGGCGCTACATACGCGCTTCGATGTCGCTGAGCGGATACATGCCCCCGTTGTGCGACCCGGTAGACGGCCACCTCCTATTGGACGGCGGTTACGTCAACAACCTGCCAGCTGACGTTATGAGGTCTTTGGGAGCGAAACACATATTGGCCATCGACGTTGGATCTCAAGATGATACAGATCTAACGAACTACGGAGACGACTTGTCGGGCTGGTGGTTGCTCTGGAAAAGATGGAACCCGTTCACGACGCCGGTCAAAGTTCCCAATCTTCCCGACATACAGAGTCGATTGGCTTACGTGTCTTGCAACAGACAGTTGGAGGAAGTGAAGAAATCGGATTACTGCGAATACATCCGTCCCCCGATCGACGCGTACAAGACGCTGCAGTTCGGCTCGTTCGACGAGATACGCGAGGTCGGCTACCGGCACGGCGCCGCCTACTTCGAGGGGCAGCGGCGCGGCGGGGGCGGGGGGGTCAGCGGGGCCGCCGCCTCCGACCGCAAGGGACCCGAGCAGCAACCGGCGCTCACCGATTATACGTTCACGGACTTAGCTCAGATGGTGTGCTCGGTCCGCACCGGTCGCGACGCCGACAACGATTCCTCCTCGTCTTCAGACTACGAGGAGGATCAGAGGCACTTCGAGGGGTACGCCAGCGAGCCCAGCGCTGGGATACTGGAGGAGGGTATAAGGGCCCGACGCGTGGGAGGGTCCCTCTCGCTGTCGGAGGACGAGGTCGACTCGGAGGCGGAGGTGTACGATCCGCTGAACAAGCGCGGCTCGAGCAGATGA
Protein
MVTQTRRSTTVMAVRDSELAKLPEGLFNAIKLRFPVVVTRLINLLGHRILGSWQKPTRSLGGAPAIDSRPLQHNFSTVAVVPVSDDVPLTAFTYELYHSLCAIGPTVRLTSDVIRKLLGLTIMDPNNEYRLSSWLAQQEDKHKVALYQCDPSLTQWTQRCIRQADCILIVALGDKQPSIGKIEKEIERLAIRTQKELVLLHREGGPNPSGTVHWLNMRTWVSQHHHVRCPHRMFTRKSQYRISELYSKVLMSEANVHSDFSRLARWLTATAVGLVLGGGGARGAAHVGMIRAIQEAGIPIDMVGGVSIGAFMGALWCMERNITTVTQKAREWSKKMTQWGRQLLDLTYPATSMFSGRQFNATIRATFGEVHIEDLWLPYFTVTTDISSSCMRIHRHGSLWRYIRASMSLSGYMPPLCDPVDGHLLLDGGYVNNLPADVMRSLGAKHILAIDVGSQDDTDLTNYGDDLSGWWLLWKRWNPFTTPVKVPNLPDIQSRLAYVSCNRQLEEVKKSDYCEYIRPPIDAYKTLQFGSFDEIREVGYRHGAAYFEGQRRGGGGGVSGAAASDRKGPEQQPALTDYTFTDLAQMVCSVRTGRDADNDSSSSSDYEEDQRHFEGYASEPSAGILEEGIRARRVGGSLSLSEDEVDSEAEVYDPLNKRGSSR
Summary
Description
Phospholipase B that deacylates intracellular phosphatidylcholine (PtdCho), generating glycerophosphocholine (GroPtdCho). This deacylation occurs at both sn-2 and sn-1 positions of PtdCho. Its specific chemical modification by certain organophosphorus (OP) compounds leads to distal axonopathy. Plays a role in the signaling mechanism between neurons and glia that regulates glia wrapping during development of the adult brain. Essential for membrane lipid homeostasis and cell survival in both neurons and glia of the adult brain.
Catalytic Activity
a 1-acyl-sn-glycero-3-phosphocholine + H2O = a fatty acid + H(+) + sn-glycerol 3-phosphocholine
Subunit
Interacts with Pka-C3; interaction inhibits the catalytic function of Pka-C3 and the esterase activity of sws.
Similarity
Belongs to the NTE family.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Endoplasmic reticulum
Hydrolase
Lipid degradation
Lipid metabolism
Membrane
Neurogenesis
Phosphoprotein
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Neuropathy target esterase sws
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JEI1
A0A3S2MAJ0
D6W9Q7
A0A026WPN4
A0A3L8DN19
E2BKL4
+ More
K7IQ10 A0A2J7QV86 A0A2J7QV89 A0A1B6CYY3 E2A0X2 A0A154PJP7 A0A067RS17 A0A158NV62 A0A0L7R5B4 A0A088ACE8 A0A195BQH0 A0A310SL74 A0A195CH99 A0A1Y1MJ22 A0A1Y1MMU8 A0A0C9RHG8 A0A1Y1MJ32 A0A151JN95 A0A1Y1MJA9 A0A1Y1MPB8 A0A151XE85 F4W4X6 A0A232F867 A0A2H1W600 A0A1B6F290 Q16I35 B0W7V2 A0A1Q3FE81 A0A023F3T3 A0A0V0G706 A0A0A9XBN8 A0A0A9XIY4 A0A224XEX9 A0A069DWW5 A0A0A1XGU3 A0A1I8M443 A0A0K8V7Q6 T1HIF7 A0A034V8P2 T1PMW0 A0A0N7Z8B7 A0A1I8P2Y4 A0A084VLT2 A0A1J1IYU6 A0A182MPR2 A0A1L8DZF0 A0A336MD57 A0A182PFX8 Q9U969 A0A0R3P339 A0A0R1EJ12 A0A0R3NY08 A0A0R3NYB8 A0A0Q5TGH0 A0A182XDV8 A0A1S4G951 A0A0Q5TKA4 A0A1W4VIK0 Q7PRV7 A0A1I8M435 A0A0Q9WTH0 A0A182U619 A0A1J1IVY4 A0A0P8XK54 A0A1I8P2X7 W8APK1 A0A182W5J3 Q9U969-3 A0A0R1EB53 E1JJF4 E0VWM1 A0A0Q9W766 A0A1W4VWL3 A0A3B0JY38 A0A0Q9XNK5 I6TQD9 A0A0P8XJV1 A0A2S2NHV6 A0A182FJG4 W5JW82 A0A2M3ZG82 A0A2M4AA44 A0A2M4A6K6 A0A2M4AIR1 A0A0P6D011 A0A0N8E5X3 A0A0P6EES6 A0A087T7D2 A0A0P5TJ89 A0A0P5VMZ6 A0A0P5I455 A0A0N8BYJ2 A0A0P4XIS7 A0A0P6BYK3
K7IQ10 A0A2J7QV86 A0A2J7QV89 A0A1B6CYY3 E2A0X2 A0A154PJP7 A0A067RS17 A0A158NV62 A0A0L7R5B4 A0A088ACE8 A0A195BQH0 A0A310SL74 A0A195CH99 A0A1Y1MJ22 A0A1Y1MMU8 A0A0C9RHG8 A0A1Y1MJ32 A0A151JN95 A0A1Y1MJA9 A0A1Y1MPB8 A0A151XE85 F4W4X6 A0A232F867 A0A2H1W600 A0A1B6F290 Q16I35 B0W7V2 A0A1Q3FE81 A0A023F3T3 A0A0V0G706 A0A0A9XBN8 A0A0A9XIY4 A0A224XEX9 A0A069DWW5 A0A0A1XGU3 A0A1I8M443 A0A0K8V7Q6 T1HIF7 A0A034V8P2 T1PMW0 A0A0N7Z8B7 A0A1I8P2Y4 A0A084VLT2 A0A1J1IYU6 A0A182MPR2 A0A1L8DZF0 A0A336MD57 A0A182PFX8 Q9U969 A0A0R3P339 A0A0R1EJ12 A0A0R3NY08 A0A0R3NYB8 A0A0Q5TGH0 A0A182XDV8 A0A1S4G951 A0A0Q5TKA4 A0A1W4VIK0 Q7PRV7 A0A1I8M435 A0A0Q9WTH0 A0A182U619 A0A1J1IVY4 A0A0P8XK54 A0A1I8P2X7 W8APK1 A0A182W5J3 Q9U969-3 A0A0R1EB53 E1JJF4 E0VWM1 A0A0Q9W766 A0A1W4VWL3 A0A3B0JY38 A0A0Q9XNK5 I6TQD9 A0A0P8XJV1 A0A2S2NHV6 A0A182FJG4 W5JW82 A0A2M3ZG82 A0A2M4AA44 A0A2M4A6K6 A0A2M4AIR1 A0A0P6D011 A0A0N8E5X3 A0A0P6EES6 A0A087T7D2 A0A0P5TJ89 A0A0P5VMZ6 A0A0P5I455 A0A0N8BYJ2 A0A0P4XIS7 A0A0P6BYK3
Pubmed
19121390
18362917
19820115
24508170
30249741
20798317
+ More
20075255 24845553 21347285 28004739 21719571 28648823 17510324 25474469 25401762 26823975 26334808 25830018 25315136 25348373 27129103 24438588 9295388 10731132 12537572 12537569 15772346 18945896 18327897 15632085 17994087 17550304 23185243 12364791 24495485 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863 20920257 23761445
20075255 24845553 21347285 28004739 21719571 28648823 17510324 25474469 25401762 26823975 26334808 25830018 25315136 25348373 27129103 24438588 9295388 10731132 12537572 12537569 15772346 18945896 18327897 15632085 17994087 17550304 23185243 12364791 24495485 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863 20920257 23761445
EMBL
BABH01003155
RSAL01000001
RVE55177.1
KQ971312
EEZ98531.2
KK107135
+ More
EZA58017.1 QOIP01000006 RLU21676.1 GL448819 EFN83712.1 NEVH01010478 PNF32498.1 PNF32499.1 GEDC01018845 GEDC01013659 JAS18453.1 JAS23639.1 GL435626 EFN73025.1 KQ434923 KZC11508.1 KK852460 KDR23520.1 ADTU01026908 ADTU01026909 ADTU01026910 KQ414654 KOC66038.1 KQ976424 KYM88209.1 KQ760132 OAD61817.1 KQ977827 KYM99458.1 GEZM01030135 GEZM01030129 JAV85792.1 GEZM01030133 JAV85795.1 GBYB01006371 JAG76138.1 GEZM01030128 JAV85802.1 KQ978850 KYN27955.1 GEZM01030136 JAV85791.1 GEZM01030138 GEZM01030132 JAV85796.1 KQ982254 KYQ58682.1 GL887596 EGI70672.1 NNAY01000723 OXU26792.1 ODYU01006561 SOQ48510.1 GECZ01025400 JAS44369.1 CH478114 EAT33921.1 DS231855 EDS38246.1 GFDL01009172 JAV25873.1 GBBI01002661 JAC16051.1 GECL01002914 JAP03210.1 GBHO01026548 GBRD01007579 JAG17056.1 JAG58242.1 GBHO01026549 GBRD01007580 GDHC01009539 JAG17055.1 JAG58241.1 JAQ09090.1 GFTR01008088 JAW08338.1 GBGD01000534 JAC88355.1 GBXI01004115 JAD10177.1 GDHF01017352 JAI34962.1 ACPB03001249 GAKP01021029 JAC37923.1 KA650107 AFP64736.1 GDKW01003795 JAI52800.1 ATLV01014551 KE524974 KFB38926.1 CVRI01000063 CRL04334.1 AXCM01003763 GFDF01002287 JAV11797.1 UFQT01000668 SSX26327.1 Z97187 AE014298 BT030819 AY061431 CH379063 KRT05973.1 CM000162 KRK06711.1 KRT05974.1 KRT05975.1 KRT05972.1 CH954180 KQS30549.1 AAAB01008846 KQS30548.1 EAA06510.6 CH963925 KRF98861.1 CRL04335.1 CH902622 KPU75112.1 GAMC01018608 JAB87947.1 KRK06712.1 BT133162 AAN09222.2 AEZ02855.1 DS235822 EEB17777.1 CH940653 KRF80723.1 OUUW01000003 SPP78276.1 CH933811 KRG06868.1 JN989963 AFM78641.1 KPU75113.1 GGMR01004120 MBY16739.1 ADMH02000256 ETN67229.1 GGFM01006826 MBW27577.1 GGFK01004291 MBW37612.1 GGFK01003106 MBW36427.1 GGFK01007342 MBW40663.1 GDIP01007524 JAM96191.1 GDIQ01058684 JAN36053.1 GDIQ01064339 JAN30398.1 KK113780 KFM61021.1 GDIP01125524 JAL78190.1 GDIP01097833 JAM05882.1 GDIQ01220540 JAK31185.1 GDIQ01132323 JAL19403.1 GDIP01240620 JAI82781.1 GDIP01007525 JAM96190.1
EZA58017.1 QOIP01000006 RLU21676.1 GL448819 EFN83712.1 NEVH01010478 PNF32498.1 PNF32499.1 GEDC01018845 GEDC01013659 JAS18453.1 JAS23639.1 GL435626 EFN73025.1 KQ434923 KZC11508.1 KK852460 KDR23520.1 ADTU01026908 ADTU01026909 ADTU01026910 KQ414654 KOC66038.1 KQ976424 KYM88209.1 KQ760132 OAD61817.1 KQ977827 KYM99458.1 GEZM01030135 GEZM01030129 JAV85792.1 GEZM01030133 JAV85795.1 GBYB01006371 JAG76138.1 GEZM01030128 JAV85802.1 KQ978850 KYN27955.1 GEZM01030136 JAV85791.1 GEZM01030138 GEZM01030132 JAV85796.1 KQ982254 KYQ58682.1 GL887596 EGI70672.1 NNAY01000723 OXU26792.1 ODYU01006561 SOQ48510.1 GECZ01025400 JAS44369.1 CH478114 EAT33921.1 DS231855 EDS38246.1 GFDL01009172 JAV25873.1 GBBI01002661 JAC16051.1 GECL01002914 JAP03210.1 GBHO01026548 GBRD01007579 JAG17056.1 JAG58242.1 GBHO01026549 GBRD01007580 GDHC01009539 JAG17055.1 JAG58241.1 JAQ09090.1 GFTR01008088 JAW08338.1 GBGD01000534 JAC88355.1 GBXI01004115 JAD10177.1 GDHF01017352 JAI34962.1 ACPB03001249 GAKP01021029 JAC37923.1 KA650107 AFP64736.1 GDKW01003795 JAI52800.1 ATLV01014551 KE524974 KFB38926.1 CVRI01000063 CRL04334.1 AXCM01003763 GFDF01002287 JAV11797.1 UFQT01000668 SSX26327.1 Z97187 AE014298 BT030819 AY061431 CH379063 KRT05973.1 CM000162 KRK06711.1 KRT05974.1 KRT05975.1 KRT05972.1 CH954180 KQS30549.1 AAAB01008846 KQS30548.1 EAA06510.6 CH963925 KRF98861.1 CRL04335.1 CH902622 KPU75112.1 GAMC01018608 JAB87947.1 KRK06712.1 BT133162 AAN09222.2 AEZ02855.1 DS235822 EEB17777.1 CH940653 KRF80723.1 OUUW01000003 SPP78276.1 CH933811 KRG06868.1 JN989963 AFM78641.1 KPU75113.1 GGMR01004120 MBY16739.1 ADMH02000256 ETN67229.1 GGFM01006826 MBW27577.1 GGFK01004291 MBW37612.1 GGFK01003106 MBW36427.1 GGFK01007342 MBW40663.1 GDIP01007524 JAM96191.1 GDIQ01058684 JAN36053.1 GDIQ01064339 JAN30398.1 KK113780 KFM61021.1 GDIP01125524 JAL78190.1 GDIP01097833 JAM05882.1 GDIQ01220540 JAK31185.1 GDIQ01132323 JAL19403.1 GDIP01240620 JAI82781.1 GDIP01007525 JAM96190.1
Proteomes
UP000005204
UP000283053
UP000007266
UP000053097
UP000279307
UP000008237
+ More
UP000002358 UP000235965 UP000000311 UP000076502 UP000027135 UP000005205 UP000053825 UP000005203 UP000078540 UP000078542 UP000078492 UP000075809 UP000007755 UP000215335 UP000008820 UP000002320 UP000095301 UP000015103 UP000095300 UP000030765 UP000183832 UP000075883 UP000075885 UP000000803 UP000001819 UP000002282 UP000008711 UP000076407 UP000192221 UP000007062 UP000007798 UP000075902 UP000007801 UP000075920 UP000009046 UP000008792 UP000268350 UP000009192 UP000069272 UP000000673 UP000054359
UP000002358 UP000235965 UP000000311 UP000076502 UP000027135 UP000005205 UP000053825 UP000005203 UP000078540 UP000078542 UP000078492 UP000075809 UP000007755 UP000215335 UP000008820 UP000002320 UP000095301 UP000015103 UP000095300 UP000030765 UP000183832 UP000075883 UP000075885 UP000000803 UP000001819 UP000002282 UP000008711 UP000076407 UP000192221 UP000007062 UP000007798 UP000075902 UP000007801 UP000075920 UP000009046 UP000008792 UP000268350 UP000009192 UP000069272 UP000000673 UP000054359
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JEI1
A0A3S2MAJ0
D6W9Q7
A0A026WPN4
A0A3L8DN19
E2BKL4
+ More
K7IQ10 A0A2J7QV86 A0A2J7QV89 A0A1B6CYY3 E2A0X2 A0A154PJP7 A0A067RS17 A0A158NV62 A0A0L7R5B4 A0A088ACE8 A0A195BQH0 A0A310SL74 A0A195CH99 A0A1Y1MJ22 A0A1Y1MMU8 A0A0C9RHG8 A0A1Y1MJ32 A0A151JN95 A0A1Y1MJA9 A0A1Y1MPB8 A0A151XE85 F4W4X6 A0A232F867 A0A2H1W600 A0A1B6F290 Q16I35 B0W7V2 A0A1Q3FE81 A0A023F3T3 A0A0V0G706 A0A0A9XBN8 A0A0A9XIY4 A0A224XEX9 A0A069DWW5 A0A0A1XGU3 A0A1I8M443 A0A0K8V7Q6 T1HIF7 A0A034V8P2 T1PMW0 A0A0N7Z8B7 A0A1I8P2Y4 A0A084VLT2 A0A1J1IYU6 A0A182MPR2 A0A1L8DZF0 A0A336MD57 A0A182PFX8 Q9U969 A0A0R3P339 A0A0R1EJ12 A0A0R3NY08 A0A0R3NYB8 A0A0Q5TGH0 A0A182XDV8 A0A1S4G951 A0A0Q5TKA4 A0A1W4VIK0 Q7PRV7 A0A1I8M435 A0A0Q9WTH0 A0A182U619 A0A1J1IVY4 A0A0P8XK54 A0A1I8P2X7 W8APK1 A0A182W5J3 Q9U969-3 A0A0R1EB53 E1JJF4 E0VWM1 A0A0Q9W766 A0A1W4VWL3 A0A3B0JY38 A0A0Q9XNK5 I6TQD9 A0A0P8XJV1 A0A2S2NHV6 A0A182FJG4 W5JW82 A0A2M3ZG82 A0A2M4AA44 A0A2M4A6K6 A0A2M4AIR1 A0A0P6D011 A0A0N8E5X3 A0A0P6EES6 A0A087T7D2 A0A0P5TJ89 A0A0P5VMZ6 A0A0P5I455 A0A0N8BYJ2 A0A0P4XIS7 A0A0P6BYK3
K7IQ10 A0A2J7QV86 A0A2J7QV89 A0A1B6CYY3 E2A0X2 A0A154PJP7 A0A067RS17 A0A158NV62 A0A0L7R5B4 A0A088ACE8 A0A195BQH0 A0A310SL74 A0A195CH99 A0A1Y1MJ22 A0A1Y1MMU8 A0A0C9RHG8 A0A1Y1MJ32 A0A151JN95 A0A1Y1MJA9 A0A1Y1MPB8 A0A151XE85 F4W4X6 A0A232F867 A0A2H1W600 A0A1B6F290 Q16I35 B0W7V2 A0A1Q3FE81 A0A023F3T3 A0A0V0G706 A0A0A9XBN8 A0A0A9XIY4 A0A224XEX9 A0A069DWW5 A0A0A1XGU3 A0A1I8M443 A0A0K8V7Q6 T1HIF7 A0A034V8P2 T1PMW0 A0A0N7Z8B7 A0A1I8P2Y4 A0A084VLT2 A0A1J1IYU6 A0A182MPR2 A0A1L8DZF0 A0A336MD57 A0A182PFX8 Q9U969 A0A0R3P339 A0A0R1EJ12 A0A0R3NY08 A0A0R3NYB8 A0A0Q5TGH0 A0A182XDV8 A0A1S4G951 A0A0Q5TKA4 A0A1W4VIK0 Q7PRV7 A0A1I8M435 A0A0Q9WTH0 A0A182U619 A0A1J1IVY4 A0A0P8XK54 A0A1I8P2X7 W8APK1 A0A182W5J3 Q9U969-3 A0A0R1EB53 E1JJF4 E0VWM1 A0A0Q9W766 A0A1W4VWL3 A0A3B0JY38 A0A0Q9XNK5 I6TQD9 A0A0P8XJV1 A0A2S2NHV6 A0A182FJG4 W5JW82 A0A2M3ZG82 A0A2M4AA44 A0A2M4A6K6 A0A2M4AIR1 A0A0P6D011 A0A0N8E5X3 A0A0P6EES6 A0A087T7D2 A0A0P5TJ89 A0A0P5VMZ6 A0A0P5I455 A0A0N8BYJ2 A0A0P4XIS7 A0A0P6BYK3
PDB
5FYA
E-value=4.29152e-13,
Score=183
Ontologies
KEGG
GO
GO:0016042
GO:0004622
GO:0046470
GO:0016021
GO:0016787
GO:0005789
GO:0034236
GO:0061024
GO:0005737
GO:0034349
GO:0072657
GO:0006643
GO:0006644
GO:2000480
GO:0051402
GO:0007272
GO:0007608
GO:0045494
GO:0005886
GO:0005783
GO:0008152
GO:0006629
GO:0007219
GO:0007275
GO:0050793
GO:0003676
GO:0016876
GO:0043039
Topology
Subcellular location
Endoplasmic reticulum membrane
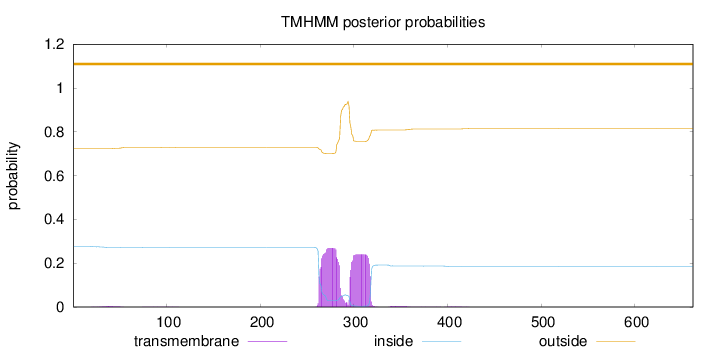
Length:
662
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.2222
Exp number, first 60 AAs:
0.08085
Total prob of N-in:
0.27593
outside
1 - 662
Population Genetic Test Statistics
Pi
279.626998
Theta
176.966218
Tajima's D
1.821376
CLR
0.562272
CSRT
0.850157492125394
Interpretation
Uncertain
