Gene
KWMTBOMO08686
Pre Gene Modal
BGIBMGA007653
Annotation
PREDICTED:_hyaluronan-mediated_motility_receptor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.575 Nuclear Reliability : 1.904
Sequence
CDS
ATGGGGACGTGTTATGAAAAACATCCATTTGAGATCGAAACTGAGGACCGTCTCACATCTATAAACACGAAAATATACGAAATCAAAAAGAAAATACAGCTGTCAGAGGGCCAAAGGAAATCTAATTACGAAGAAAACGAAGCCGAGAAACAGCAGAACAATGATCTAATAGAGGCGCTGAAGAGGGAGATAAAGTCACGTGTCGCCGAACTGATACAAGCTAGATCAGCTATAGGAGAAGACGAAGTACATGTCAAGAAGTACTTGAATGACATATGTCCTATTGGAGACAAAACAGCGGATCAGATAGCCCATAATTTAGATCTAAAAGTCATTGAACAGCGTAAAATGTTGGATCTACTGAAATACGAGAGACGCAGTAAAAAGAGGAGACTCACGGAACTGGAGAAAGAATACGAAAGCCTCTTGATTGAGAACAGCAAAGGTGAACTTGTCAAATCGAAAATTTCTAGTCGCGCCAGAAAGCTCGTTTCGCATTTAGAGAACGAAATTCATAAAGTGTTAATACAATGGACAGAAGCGGAACTGGTCAAGAAAAAGTACTTTTCTATCAGACAAGCCCTCATTGATGACTCAGTCAAATTCGAAAGTTCCTTGTCACGTATTGAAGATCTATTGAACAAACAACGAGATGAAATTAAAAAACTTGAGTATGTACGGGAAGAGGCCGCGGAGATGCGTATCCGCGCGACAGCGGCTACGGCCGCGGAGGCTGCACGTGCTCACGATGCAGAACACGCACGAGCTACGGAAAGAGCCTACTTCTCTCAAAGATTCGGCGAGAGAAAACGGGAACTTGAGAAGTTGGAGAAAAGAGTATTTCCGCCCAACAGTCGATCAGTGGTTCGTCAAGAGTCTACCAGATCAGATCGGGAAGCCGAAGCGGTATCCGAGGAAGCATCAGCCGCAGCTCAGATGGAAGACATTTTCCAGAAACTTATGAAATTAACTGGTGTCACAAATCCTGAAGAAGTGTTCGACAGGTTCCGCGCCCAGCGCGACACCTGCAAACGACTGAGCTACCTCCAGCAGACCACCGAAGAAGAGAAACTGGACCTCGAGCAGAGACAAACGACGCTGATGGCCGAACTGGAAGCTTTCAAATTCGCTTCAGTCAAAGACAAAGATGAGGGTCAAGATCAAATCAACGACTTAAAGATCGAAATACAGAAAGAGCAGGATATTCACGATGAACTACAGAAGAAAATAGACGATCTCGAAACATTTCTACTCGAAATCAAGAGATTGCTCCACGACCTCTGCAAGTTATTAGATTTGATACAGGAGCCGTCAGTGCCAGAATGGACAGCAGAAGCGCGCGACGTCCATGAATTGGTGGTGGTGCTCACATCTAGATACGAAAAAGCCAAAGCTAGATCTGATGAGATCAAGGAGAAGAATAAGGATGTTACTACGATCATAGTGCCTTCTAATCAAACTAGCGCGGCCGCTTCCGTGGCTGGACTCAGCGTGCACAGTCATAGCACGCCTAAGGAGTCTGACAAGCAAATACCAACCTATAAGGAGCTCTTGCAAAAGGAACCTGTCAGAGCACCACCATCCGATGAAGAGGAAGACATCCCGTCAAGATGCTATCTGAAGAGGCAAGCTCAAGTTATTGTCGATACCAAATGCAGAAGAAAAGGATTCCGTCCATTCCAGAGGCGATAA
Protein
MGTCYEKHPFEIETEDRLTSINTKIYEIKKKIQLSEGQRKSNYEENEAEKQQNNDLIEALKREIKSRVAELIQARSAIGEDEVHVKKYLNDICPIGDKTADQIAHNLDLKVIEQRKMLDLLKYERRSKKRRLTELEKEYESLLIENSKGELVKSKISSRARKLVSHLENEIHKVLIQWTEAELVKKKYFSIRQALIDDSVKFESSLSRIEDLLNKQRDEIKKLEYVREEAAEMRIRATAATAAEAARAHDAEHARATERAYFSQRFGERKRELEKLEKRVFPPNSRSVVRQESTRSDREAEAVSEEASAAAQMEDIFQKLMKLTGVTNPEEVFDRFRAQRDTCKRLSYLQQTTEEEKLDLEQRQTTLMAELEAFKFASVKDKDEGQDQINDLKIEIQKEQDIHDELQKKIDDLETFLLEIKRLLHDLCKLLDLIQEPSVPEWTAEARDVHELVVVLTSRYEKAKARSDEIKEKNKDVTTIIVPSNQTSAAASVAGLSVHSHSTPKESDKQIPTYKELLQKEPVRAPPSDEEEDIPSRCYLKRQAQVIVDTKCRRKGFRPFQRR
Summary
Uniprot
H9JDQ6
A0A2H1W838
A0A437BYS3
A0A2A4JKM5
A0A0L7L5J4
A0A194RF59
+ More
A0A194PR27 A0A212FGX4 A0A2W1BL79 A0A2J7PJ25 A0A1S4FPK8 A0A1I8PWU3 W5JTZ2 A0A0M4F004 A0A182GPU7 A0A2J7PJ36 A0A084WPZ6 A0A182FIW3 B0WGH7 A0A182QYE1 A0A336K8U2 B0W929 B4LF64 B4KVJ5 B3NH27 B4PFS1 B4QQP7 B3M4L7 Q9VTQ3 A0A3B0JR79 B4HF14 A0A2P8YLM8 Q2M0Z7 A0A1W4VUG1 B4GQW1 B4N3Z0 A0A182Y4Z4 A0A1B0BRE0 A0A1A9UVP5 A0A1A9YC33 A0A1B0AH18 A0A1B0G0U0 A0A0J9RU14 K7IZP1 Q16TK8 A0A182JEZ6 A0A182IDT3 A0A182PI35 A0A182NGH8 A0A182LID2 Q7QHY2 B4J3B1 A0A1A9X2X9 A0A1J1IFA4 A0A1B0D3V6 A0A2J7PJ08 X1XGR6 A0A3L8DHM1 A0A232EM43 A0A2S2PIR3 A0A3P8Z9K1
A0A194PR27 A0A212FGX4 A0A2W1BL79 A0A2J7PJ25 A0A1S4FPK8 A0A1I8PWU3 W5JTZ2 A0A0M4F004 A0A182GPU7 A0A2J7PJ36 A0A084WPZ6 A0A182FIW3 B0WGH7 A0A182QYE1 A0A336K8U2 B0W929 B4LF64 B4KVJ5 B3NH27 B4PFS1 B4QQP7 B3M4L7 Q9VTQ3 A0A3B0JR79 B4HF14 A0A2P8YLM8 Q2M0Z7 A0A1W4VUG1 B4GQW1 B4N3Z0 A0A182Y4Z4 A0A1B0BRE0 A0A1A9UVP5 A0A1A9YC33 A0A1B0AH18 A0A1B0G0U0 A0A0J9RU14 K7IZP1 Q16TK8 A0A182JEZ6 A0A182IDT3 A0A182PI35 A0A182NGH8 A0A182LID2 Q7QHY2 B4J3B1 A0A1A9X2X9 A0A1J1IFA4 A0A1B0D3V6 A0A2J7PJ08 X1XGR6 A0A3L8DHM1 A0A232EM43 A0A2S2PIR3 A0A3P8Z9K1
Pubmed
EMBL
BABH01003155
ODYU01006636
SOQ48654.1
RSAL01000001
RVE55179.1
NWSH01001118
+ More
PCG72511.1 JTDY01002873 KOB70564.1 KQ460473 KPJ14561.1 KQ459596 KPI95413.1 AGBW02008589 OWR52982.1 KZ150050 PZC74425.1 NEVH01024952 PNF16324.1 ADMH02000382 ETN66763.1 CP012525 ALC44387.1 ALC44484.1 JXUM01001860 KQ560129 KXJ84291.1 PNF16323.1 ATLV01025144 KE525369 KFB52290.1 DS231926 EDS26974.1 AXCN02002557 UFQS01000180 UFQT01000180 SSX00827.1 SSX21207.1 DS231861 EDS39466.1 CH940647 EDW70252.1 CH933809 EDW19466.1 CH954178 EDV51484.1 CM000159 EDW94220.1 CM000363 EDX10149.1 CH902618 EDV39416.1 AE014296 AAF49993.1 OUUW01000009 SPP84667.1 CH480815 EDW41183.1 PYGN01000508 PSN45156.1 CH379069 EAL30779.1 CH479188 EDW40146.1 CH964095 EDW79345.1 JXJN01019051 CCAG010021774 CM002912 KMY99107.1 CH477646 EAT37842.1 APCN01004793 AAAB01008811 EAA04963.5 CH916366 EDV97210.1 CVRI01000048 CRK98896.1 AJVK01011049 AJVK01011050 AJVK01011051 PNF16322.1 ABLF02023729 QOIP01000008 RLU19917.1 NNAY01003448 OXU19401.1 GGMR01016678 MBY29297.1
PCG72511.1 JTDY01002873 KOB70564.1 KQ460473 KPJ14561.1 KQ459596 KPI95413.1 AGBW02008589 OWR52982.1 KZ150050 PZC74425.1 NEVH01024952 PNF16324.1 ADMH02000382 ETN66763.1 CP012525 ALC44387.1 ALC44484.1 JXUM01001860 KQ560129 KXJ84291.1 PNF16323.1 ATLV01025144 KE525369 KFB52290.1 DS231926 EDS26974.1 AXCN02002557 UFQS01000180 UFQT01000180 SSX00827.1 SSX21207.1 DS231861 EDS39466.1 CH940647 EDW70252.1 CH933809 EDW19466.1 CH954178 EDV51484.1 CM000159 EDW94220.1 CM000363 EDX10149.1 CH902618 EDV39416.1 AE014296 AAF49993.1 OUUW01000009 SPP84667.1 CH480815 EDW41183.1 PYGN01000508 PSN45156.1 CH379069 EAL30779.1 CH479188 EDW40146.1 CH964095 EDW79345.1 JXJN01019051 CCAG010021774 CM002912 KMY99107.1 CH477646 EAT37842.1 APCN01004793 AAAB01008811 EAA04963.5 CH916366 EDV97210.1 CVRI01000048 CRK98896.1 AJVK01011049 AJVK01011050 AJVK01011051 PNF16322.1 ABLF02023729 QOIP01000008 RLU19917.1 NNAY01003448 OXU19401.1 GGMR01016678 MBY29297.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000037510
UP000053240
UP000053268
+ More
UP000007151 UP000235965 UP000095300 UP000000673 UP000092553 UP000069940 UP000249989 UP000030765 UP000069272 UP000002320 UP000075886 UP000008792 UP000009192 UP000008711 UP000002282 UP000000304 UP000007801 UP000000803 UP000268350 UP000001292 UP000245037 UP000001819 UP000192221 UP000008744 UP000007798 UP000076408 UP000092460 UP000078200 UP000092443 UP000092445 UP000092444 UP000002358 UP000008820 UP000075880 UP000075840 UP000075885 UP000075884 UP000075882 UP000007062 UP000001070 UP000091820 UP000183832 UP000092462 UP000007819 UP000279307 UP000215335 UP000265140
UP000007151 UP000235965 UP000095300 UP000000673 UP000092553 UP000069940 UP000249989 UP000030765 UP000069272 UP000002320 UP000075886 UP000008792 UP000009192 UP000008711 UP000002282 UP000000304 UP000007801 UP000000803 UP000268350 UP000001292 UP000245037 UP000001819 UP000192221 UP000008744 UP000007798 UP000076408 UP000092460 UP000078200 UP000092443 UP000092445 UP000092444 UP000002358 UP000008820 UP000075880 UP000075840 UP000075885 UP000075884 UP000075882 UP000007062 UP000001070 UP000091820 UP000183832 UP000092462 UP000007819 UP000279307 UP000215335 UP000265140
Interpro
Gene 3D
ProteinModelPortal
H9JDQ6
A0A2H1W838
A0A437BYS3
A0A2A4JKM5
A0A0L7L5J4
A0A194RF59
+ More
A0A194PR27 A0A212FGX4 A0A2W1BL79 A0A2J7PJ25 A0A1S4FPK8 A0A1I8PWU3 W5JTZ2 A0A0M4F004 A0A182GPU7 A0A2J7PJ36 A0A084WPZ6 A0A182FIW3 B0WGH7 A0A182QYE1 A0A336K8U2 B0W929 B4LF64 B4KVJ5 B3NH27 B4PFS1 B4QQP7 B3M4L7 Q9VTQ3 A0A3B0JR79 B4HF14 A0A2P8YLM8 Q2M0Z7 A0A1W4VUG1 B4GQW1 B4N3Z0 A0A182Y4Z4 A0A1B0BRE0 A0A1A9UVP5 A0A1A9YC33 A0A1B0AH18 A0A1B0G0U0 A0A0J9RU14 K7IZP1 Q16TK8 A0A182JEZ6 A0A182IDT3 A0A182PI35 A0A182NGH8 A0A182LID2 Q7QHY2 B4J3B1 A0A1A9X2X9 A0A1J1IFA4 A0A1B0D3V6 A0A2J7PJ08 X1XGR6 A0A3L8DHM1 A0A232EM43 A0A2S2PIR3 A0A3P8Z9K1
A0A194PR27 A0A212FGX4 A0A2W1BL79 A0A2J7PJ25 A0A1S4FPK8 A0A1I8PWU3 W5JTZ2 A0A0M4F004 A0A182GPU7 A0A2J7PJ36 A0A084WPZ6 A0A182FIW3 B0WGH7 A0A182QYE1 A0A336K8U2 B0W929 B4LF64 B4KVJ5 B3NH27 B4PFS1 B4QQP7 B3M4L7 Q9VTQ3 A0A3B0JR79 B4HF14 A0A2P8YLM8 Q2M0Z7 A0A1W4VUG1 B4GQW1 B4N3Z0 A0A182Y4Z4 A0A1B0BRE0 A0A1A9UVP5 A0A1A9YC33 A0A1B0AH18 A0A1B0G0U0 A0A0J9RU14 K7IZP1 Q16TK8 A0A182JEZ6 A0A182IDT3 A0A182PI35 A0A182NGH8 A0A182LID2 Q7QHY2 B4J3B1 A0A1A9X2X9 A0A1J1IFA4 A0A1B0D3V6 A0A2J7PJ08 X1XGR6 A0A3L8DHM1 A0A232EM43 A0A2S2PIR3 A0A3P8Z9K1
Ontologies
GO
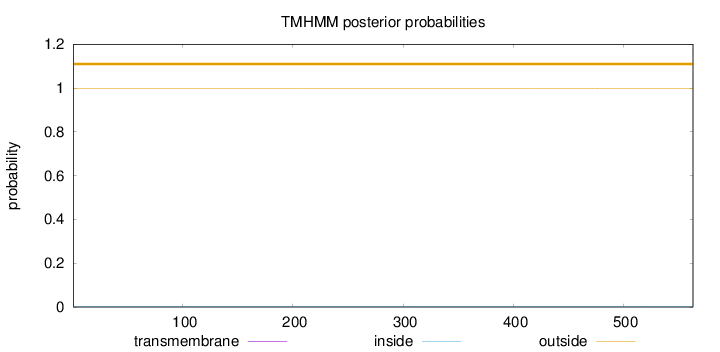
Topology
Length:
563
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00084
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00174
outside
1 - 563
Population Genetic Test Statistics
Pi
258.615313
Theta
178.303881
Tajima's D
1.069954
CLR
0.298804
CSRT
0.681065946702665
Interpretation
Uncertain
