Gene
KWMTBOMO08684
Pre Gene Modal
BGIBMGA007930
Annotation
PREDICTED:_tyramine_receptor_Ser-2-like_[Amyelois_transitella]
Full name
5-hydroxytryptamine receptor 1D
+ More
Alpha-1A adrenergic receptor
Alpha-1A adrenergic receptor
Alternative Name
Serotonin receptor 1D
Alpha-1A adrenoreceptor
MAR1
Alpha-1A adrenoreceptor
MAR1
Location in the cell
PlasmaMembrane Reliability : 4.64
Sequence
CDS
ATGCAAAGTGTGTTGACGCTCCACCCTCGCGGTGACATGACGAGCTCGTGGTGGGCGGCTGCGGCAGCTATGGCCGCGGTGACTTTCTTCACGGTCGGTGGGAACGCTGCTGTGCTGGTGGCGCTGAAGAGAGTTAATCGCGCCCCAGCACACTATCCCTTGGCGAGCTTAGCGACCGCAGACTTGCTTCTCGGTCTACTAGTGCTGCCAATAGCAGCCGCCAGGGAATTGTTCGTCTTTCATTTGAACTGGGCAATATGTTCCTGTTGGAGCACCTTGGATGTCCTATGCTGCACCGCCTCTATCCTCTCCATCTGCACCCTAGGGTGGGAGAGATTCTGCGGCATCACCGCTCCGTTAGCTCGTGCACAGAGAGCAAAGACCGCTAGACTCTTGGCTGCACTGATCTGGCCCGTGGCGACAACAATCGCTATACCGACTGCCTTCATTCCTTCGCCGAAGCATTTTCTACCTGGAGAAATAGCTAAGGCTTGTACTGTTAACACTAATTCTAGTTACGTATTCGTCAGTATTTTCTTTTCGTTTTATCTTCCCGCAAGTGTGATGGTTGTTCTTTATGCCCGGATACTGCGAGCTCTGGCGGCACCGCCCTCAATCAGGTTCCATCGTGGCCAGGTCACAAACAATGTTGAAAAAGTCATCCCCGATGCTGTCGTGGTAAATGTGGACAGCGTTACTCCTGCAGATGAATCACAAGAGAATCATAAAGTACCCAAGAGCATAGTGGTACCACCTTCTCCAAAAACCGGCCATTTCATGATACCCTCGTCGGAAACGGTACGTGCCATAACTTCTCCTTTAAAGAGACTCAAATTTCCGGTCGCGGGTATAATATCGCGCCAACGTCGTGCCACCCGGACCATCGTGATGCTGATGTCACTGTTCCTACTGTGCTGGACGCCGTTCTTCGTGATGCTTCCCGTAGATTCTATATGCGAGTGCGTCAGAGATGCCGCGTGGTTGTGGTGCACTTGGCTTGGGTACACGAACTCGGCTCTCAATCCGCTGGTCTACGCGGCCTCCTCCCCGGCGGTCAGTCGGGCCCTCCACGCCTCCCTCACCACCACTACATGCACCAGAACTACGGATGTTACTCGTACTCCTTTACCGAGAAAATAG
Protein
MQSVLTLHPRGDMTSSWWAAAAAMAAVTFFTVGGNAAVLVALKRVNRAPAHYPLASLATADLLLGLLVLPIAAARELFVFHLNWAICSCWSTLDVLCCTASILSICTLGWERFCGITAPLARAQRAKTARLLAALIWPVATTIAIPTAFIPSPKHFLPGEIAKACTVNTNSSYVFVSIFFSFYLPASVMVVLYARILRALAAPPSIRFHRGQVTNNVEKVIPDAVVVNVDSVTPADESQENHKVPKSIVVPPSPKTGHFMIPSSETVRAITSPLKRLKFPVAGIISRQRRATRTIVMLMSLFLLCWTPFFVMLPVDSICECVRDAAWLWCTWLGYTNSALNPLVYAASSPAVSRALHASLTTTTCTRTTDVTRTPLPRK
Summary
Description
G-protein coupled receptor for 5-hydroxytryptamine (serotonin). Also functions as a receptor for various alkaloids and psychoactive substances. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors, such as adenylate cyclase. Signaling inhibits adenylate cyclase activity. Regulates the release of 5-hydroxytryptamine in the brain, and thereby affects neural activity. May also play a role in regulating the release of other neurotransmitters. May play a role in vasoconstriction (By similarity).
This alpha-adrenergic receptor mediates its action by association with G proteins that activate a phosphatidylinositol-calcium second messenger system.
G-protein coupled receptor for 5-hydroxytryptamine (serotonin). Also functions as a receptor for various alkaloids and psychoactive substances. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors, such as adenylate cyclase. Signaling inhibits adenylate cyclase activity. Regulates the release of 5-hydroxytryptamine in the brain, and thereby affects neural activity. May also play a role in regulating the release of other neurotransmitters. May play a role in vasoconstriction.
This alpha-adrenergic receptor mediates its action by association with G proteins that activate a phosphatidylinositol-calcium second messenger system.
G-protein coupled receptor for 5-hydroxytryptamine (serotonin). Also functions as a receptor for various alkaloids and psychoactive substances. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors, such as adenylate cyclase. Signaling inhibits adenylate cyclase activity. Regulates the release of 5-hydroxytryptamine in the brain, and thereby affects neural activity. May also play a role in regulating the release of other neurotransmitters. May play a role in vasoconstriction.
Subunit
Homodimer. Heterodimer with HTR1B (By similarity).
Similarity
Belongs to the G-protein coupled receptor 1 family.
Belongs to the G-protein coupled receptor 1 family. Adrenergic receptor subfamily. ADRA1A sub-subfamily.
Belongs to the G-protein coupled receptor 1 family. Adrenergic receptor subfamily. ADRA1A sub-subfamily.
Keywords
Cell membrane
Complete proteome
Disulfide bond
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Lipoprotein
Palmitate
Phosphoprotein
Feature
chain 5-hydroxytryptamine receptor 1D
Uniprot
H9JEI3
A0A437BY54
A0A212EGU3
A0A2H1VAC3
A0A2W1BRP0
A0A194RF64
+ More
A0A194PWA1 A0A091RZT6 U3I2H0 R0KN79 A0A182FPD0 A0A091I5X8 U3KIP3 A0A2K5XR79 A0A2K5HFW3 A0A2K6AWB4 A0A2K5UMI0 G7MHG5 A0A091EPK7 A0A2M3Z755 A0A2M3Z6T3 A0A096MXA4 C3Y3T3 F7FW05 A0A0D9S925 A0A2K5BZX2 A0A2K6S3B0 I3M8J5 A0A2K5PAY1 A0A2K5KLB4 G7NVU4 A0A218UXY1 H2ZTR3 P79400 H0YT67 I3L844 L5JV21 Q9N263 S7PXM8 A0A2D0QXV0 F6V9Y1 A0A2M4BJ57 A0A2M4BJY8 A0A2M4BJ77 A0A2M4BJ74 A0A383ZJX1 A0A2M4AM59 A0A1S3A3K6 A0A2M4CPW4 A0A060VTR1 G1Q9M2 Q2VA56 A0A3B1J4P6 W5QAN1 G1KD63 F7AXV1 A0A3Q3BZI2 A0A3B3DEV3 A0A3Q0QU85 A0A437D146 A0A3Q7MZ24 W5JAC2 A0A3P4S279 A0A2U3VIY5 A0A452E6Y0 Q91175 A0A383YYT4 G3TQH0 A0A3Q4N0A6 A0A0U2M149 A0A3B4BRC6 A0A1S3NJV2 A0A1U7TMX9 A0A341CF30 W5NP23 A0A2Y9PLB4 H2M9N5 A0A3Q2VFL3 A0A3P8PFR7 A0A2Y9DMB8 A0A3P9LSQ7 X2C6U2 A0A3P9BWW8 A0A3B4GKQ9 A0A1S3GJD4 A0A3P9JP07 H0ZQU1 A0A3M0KJA4 A0A2P4SMU4 A0A140TAV9 A0A2K5XHR2 A0A2K5KXA7 A0A2F0B829 X2CCR5 A0A093PMC8 Q60484
A0A194PWA1 A0A091RZT6 U3I2H0 R0KN79 A0A182FPD0 A0A091I5X8 U3KIP3 A0A2K5XR79 A0A2K5HFW3 A0A2K6AWB4 A0A2K5UMI0 G7MHG5 A0A091EPK7 A0A2M3Z755 A0A2M3Z6T3 A0A096MXA4 C3Y3T3 F7FW05 A0A0D9S925 A0A2K5BZX2 A0A2K6S3B0 I3M8J5 A0A2K5PAY1 A0A2K5KLB4 G7NVU4 A0A218UXY1 H2ZTR3 P79400 H0YT67 I3L844 L5JV21 Q9N263 S7PXM8 A0A2D0QXV0 F6V9Y1 A0A2M4BJ57 A0A2M4BJY8 A0A2M4BJ77 A0A2M4BJ74 A0A383ZJX1 A0A2M4AM59 A0A1S3A3K6 A0A2M4CPW4 A0A060VTR1 G1Q9M2 Q2VA56 A0A3B1J4P6 W5QAN1 G1KD63 F7AXV1 A0A3Q3BZI2 A0A3B3DEV3 A0A3Q0QU85 A0A437D146 A0A3Q7MZ24 W5JAC2 A0A3P4S279 A0A2U3VIY5 A0A452E6Y0 Q91175 A0A383YYT4 G3TQH0 A0A3Q4N0A6 A0A0U2M149 A0A3B4BRC6 A0A1S3NJV2 A0A1U7TMX9 A0A341CF30 W5NP23 A0A2Y9PLB4 H2M9N5 A0A3Q2VFL3 A0A3P8PFR7 A0A2Y9DMB8 A0A3P9LSQ7 X2C6U2 A0A3P9BWW8 A0A3B4GKQ9 A0A1S3GJD4 A0A3P9JP07 H0ZQU1 A0A3M0KJA4 A0A2P4SMU4 A0A140TAV9 A0A2K5XHR2 A0A2K5KXA7 A0A2F0B829 X2CCR5 A0A093PMC8 Q60484
Pubmed
EMBL
BABH01003169
BABH01003170
RSAL01000001
RVE55181.1
AGBW02015022
OWR40696.1
+ More
ODYU01001500 SOQ37795.1 KZ150050 PZC74423.1 KQ460473 KPJ14566.1 KQ459596 KPI95410.1 KK938254 KFQ48619.1 ADON01133310 KB744656 EOA94663.1 KL218157 KFP02853.1 AGTO01000739 AQIA01005394 CM001253 EHH14435.1 KK718726 KFO58254.1 GGFM01003601 MBW24352.1 GGFM01003464 MBW24215.1 AHZZ02015472 GG666484 EEN64959.1 JSUE03000651 AQIB01141981 AGTP01007529 CM001276 EHH49638.1 MUZQ01000093 OWK58675.1 AFYH01257626 Y11868 ABQF01050233 AEMK02000045 DQIR01026014 DQIR01177805 HCZ81489.1 KB031148 ELK02158.1 AF117655 AAF61808.1 KE163683 EPQ13322.1 GAMT01007661 GAMS01008987 JAB04200.1 JAB14149.1 GGFJ01003965 MBW53106.1 GGFJ01003967 MBW53108.1 GGFJ01003966 MBW53107.1 GGFJ01003964 MBW53105.1 GGFK01008533 MBW41854.1 GGFL01002750 MBW66928.1 FR904262 CDQ56384.1 AAPE02027924 DQ278449 ABB83642.1 AMGL01057131 CM012445 RVE68482.1 ADMH02001946 ETN60373.1 CYRY02045770 VCX41304.1 LWLT01000002 D63859 KU052847 ALR88680.1 AHAT01009586 KC862488 KC862489 AGN73583.1 AGN73584.1 ABQF01000819 QRBI01000106 RMC13339.1 PPHD01034311 POI25433.1 AAGW02018996 NTJE010098057 MBV99163.1 KC862445 KC862446 KC862449 KC862450 KC862451 KC862454 AGN73540.1 AGN73541.1 AGN73544.1 AGN73545.1 AGN73546.1 AGN73549.1 KL670077 KFW77988.1 X94436 U82174
ODYU01001500 SOQ37795.1 KZ150050 PZC74423.1 KQ460473 KPJ14566.1 KQ459596 KPI95410.1 KK938254 KFQ48619.1 ADON01133310 KB744656 EOA94663.1 KL218157 KFP02853.1 AGTO01000739 AQIA01005394 CM001253 EHH14435.1 KK718726 KFO58254.1 GGFM01003601 MBW24352.1 GGFM01003464 MBW24215.1 AHZZ02015472 GG666484 EEN64959.1 JSUE03000651 AQIB01141981 AGTP01007529 CM001276 EHH49638.1 MUZQ01000093 OWK58675.1 AFYH01257626 Y11868 ABQF01050233 AEMK02000045 DQIR01026014 DQIR01177805 HCZ81489.1 KB031148 ELK02158.1 AF117655 AAF61808.1 KE163683 EPQ13322.1 GAMT01007661 GAMS01008987 JAB04200.1 JAB14149.1 GGFJ01003965 MBW53106.1 GGFJ01003967 MBW53108.1 GGFJ01003966 MBW53107.1 GGFJ01003964 MBW53105.1 GGFK01008533 MBW41854.1 GGFL01002750 MBW66928.1 FR904262 CDQ56384.1 AAPE02027924 DQ278449 ABB83642.1 AMGL01057131 CM012445 RVE68482.1 ADMH02001946 ETN60373.1 CYRY02045770 VCX41304.1 LWLT01000002 D63859 KU052847 ALR88680.1 AHAT01009586 KC862488 KC862489 AGN73583.1 AGN73584.1 ABQF01000819 QRBI01000106 RMC13339.1 PPHD01034311 POI25433.1 AAGW02018996 NTJE010098057 MBV99163.1 KC862445 KC862446 KC862449 KC862450 KC862451 KC862454 AGN73540.1 AGN73541.1 AGN73544.1 AGN73545.1 AGN73546.1 AGN73549.1 KL670077 KFW77988.1 X94436 U82174
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000053268
UP000016666
+ More
UP000069272 UP000054308 UP000016665 UP000233140 UP000233080 UP000233120 UP000233100 UP000052976 UP000028761 UP000001554 UP000006718 UP000029965 UP000233020 UP000233220 UP000005215 UP000233040 UP000233060 UP000009130 UP000197619 UP000008672 UP000008227 UP000007754 UP000010552 UP000221080 UP000008225 UP000261681 UP000079721 UP000193380 UP000001074 UP000018467 UP000002356 UP000001646 UP000002281 UP000264840 UP000261560 UP000261340 UP000286641 UP000000673 UP000245340 UP000291000 UP000001038 UP000265180 UP000265200 UP000007646 UP000261580 UP000261440 UP000087266 UP000189704 UP000252040 UP000018468 UP000248483 UP000265100 UP000248480 UP000265160 UP000261460 UP000081671 UP000269221 UP000001811 UP000053258 UP000005447
UP000069272 UP000054308 UP000016665 UP000233140 UP000233080 UP000233120 UP000233100 UP000052976 UP000028761 UP000001554 UP000006718 UP000029965 UP000233020 UP000233220 UP000005215 UP000233040 UP000233060 UP000009130 UP000197619 UP000008672 UP000008227 UP000007754 UP000010552 UP000221080 UP000008225 UP000261681 UP000079721 UP000193380 UP000001074 UP000018467 UP000002356 UP000001646 UP000002281 UP000264840 UP000261560 UP000261340 UP000286641 UP000000673 UP000245340 UP000291000 UP000001038 UP000265180 UP000265200 UP000007646 UP000261580 UP000261440 UP000087266 UP000189704 UP000252040 UP000018468 UP000248483 UP000265100 UP000248480 UP000265160 UP000261460 UP000081671 UP000269221 UP000001811 UP000053258 UP000005447
Pfam
PF00001 7tm_1
Interpro
ProteinModelPortal
H9JEI3
A0A437BY54
A0A212EGU3
A0A2H1VAC3
A0A2W1BRP0
A0A194RF64
+ More
A0A194PWA1 A0A091RZT6 U3I2H0 R0KN79 A0A182FPD0 A0A091I5X8 U3KIP3 A0A2K5XR79 A0A2K5HFW3 A0A2K6AWB4 A0A2K5UMI0 G7MHG5 A0A091EPK7 A0A2M3Z755 A0A2M3Z6T3 A0A096MXA4 C3Y3T3 F7FW05 A0A0D9S925 A0A2K5BZX2 A0A2K6S3B0 I3M8J5 A0A2K5PAY1 A0A2K5KLB4 G7NVU4 A0A218UXY1 H2ZTR3 P79400 H0YT67 I3L844 L5JV21 Q9N263 S7PXM8 A0A2D0QXV0 F6V9Y1 A0A2M4BJ57 A0A2M4BJY8 A0A2M4BJ77 A0A2M4BJ74 A0A383ZJX1 A0A2M4AM59 A0A1S3A3K6 A0A2M4CPW4 A0A060VTR1 G1Q9M2 Q2VA56 A0A3B1J4P6 W5QAN1 G1KD63 F7AXV1 A0A3Q3BZI2 A0A3B3DEV3 A0A3Q0QU85 A0A437D146 A0A3Q7MZ24 W5JAC2 A0A3P4S279 A0A2U3VIY5 A0A452E6Y0 Q91175 A0A383YYT4 G3TQH0 A0A3Q4N0A6 A0A0U2M149 A0A3B4BRC6 A0A1S3NJV2 A0A1U7TMX9 A0A341CF30 W5NP23 A0A2Y9PLB4 H2M9N5 A0A3Q2VFL3 A0A3P8PFR7 A0A2Y9DMB8 A0A3P9LSQ7 X2C6U2 A0A3P9BWW8 A0A3B4GKQ9 A0A1S3GJD4 A0A3P9JP07 H0ZQU1 A0A3M0KJA4 A0A2P4SMU4 A0A140TAV9 A0A2K5XHR2 A0A2K5KXA7 A0A2F0B829 X2CCR5 A0A093PMC8 Q60484
A0A194PWA1 A0A091RZT6 U3I2H0 R0KN79 A0A182FPD0 A0A091I5X8 U3KIP3 A0A2K5XR79 A0A2K5HFW3 A0A2K6AWB4 A0A2K5UMI0 G7MHG5 A0A091EPK7 A0A2M3Z755 A0A2M3Z6T3 A0A096MXA4 C3Y3T3 F7FW05 A0A0D9S925 A0A2K5BZX2 A0A2K6S3B0 I3M8J5 A0A2K5PAY1 A0A2K5KLB4 G7NVU4 A0A218UXY1 H2ZTR3 P79400 H0YT67 I3L844 L5JV21 Q9N263 S7PXM8 A0A2D0QXV0 F6V9Y1 A0A2M4BJ57 A0A2M4BJY8 A0A2M4BJ77 A0A2M4BJ74 A0A383ZJX1 A0A2M4AM59 A0A1S3A3K6 A0A2M4CPW4 A0A060VTR1 G1Q9M2 Q2VA56 A0A3B1J4P6 W5QAN1 G1KD63 F7AXV1 A0A3Q3BZI2 A0A3B3DEV3 A0A3Q0QU85 A0A437D146 A0A3Q7MZ24 W5JAC2 A0A3P4S279 A0A2U3VIY5 A0A452E6Y0 Q91175 A0A383YYT4 G3TQH0 A0A3Q4N0A6 A0A0U2M149 A0A3B4BRC6 A0A1S3NJV2 A0A1U7TMX9 A0A341CF30 W5NP23 A0A2Y9PLB4 H2M9N5 A0A3Q2VFL3 A0A3P8PFR7 A0A2Y9DMB8 A0A3P9LSQ7 X2C6U2 A0A3P9BWW8 A0A3B4GKQ9 A0A1S3GJD4 A0A3P9JP07 H0ZQU1 A0A3M0KJA4 A0A2P4SMU4 A0A140TAV9 A0A2K5XHR2 A0A2K5KXA7 A0A2F0B829 X2CCR5 A0A093PMC8 Q60484
PDB
6G79
E-value=5.0294e-20,
Score=240
Ontologies
GO
GO:0004930
GO:0016021
GO:0006939
GO:0007268
GO:0040012
GO:0042310
GO:0005887
GO:0004993
GO:0050795
GO:0051378
GO:0009636
GO:0007193
GO:0014827
GO:0007204
GO:0007188
GO:0045987
GO:0004935
GO:0001994
GO:0045907
GO:0007200
GO:0004937
GO:0071880
GO:0030425
GO:0030594
GO:0007187
GO:0005886
GO:0046849
GO:0035690
GO:0005737
GO:0071312
GO:0099154
GO:1904707
GO:0007205
GO:0099056
GO:0014063
GO:0008144
GO:0002031
GO:0007198
GO:0071502
GO:0007186
GO:0006189
GO:0005840
GO:0006426
GO:0003676
GO:0016876
GO:0043039
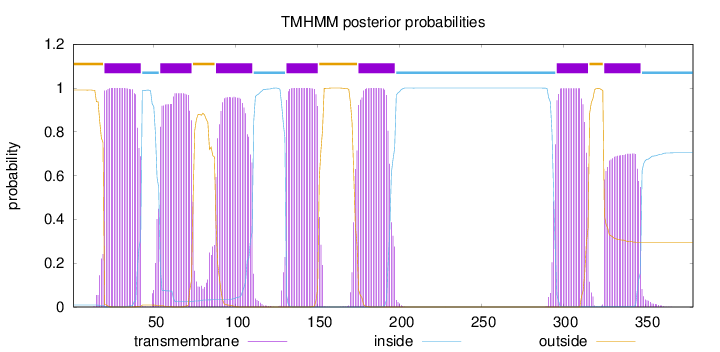
Topology
Subcellular location
Cell membrane
Length:
379
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
145.65134
Exp number, first 60 AAs:
30.40836
Total prob of N-in:
0.00926
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 53
TMhelix
54 - 73
outside
74 - 87
TMhelix
88 - 110
inside
111 - 130
TMhelix
131 - 150
outside
151 - 174
TMhelix
175 - 197
inside
198 - 295
TMhelix
296 - 315
outside
316 - 324
TMhelix
325 - 347
inside
348 - 379
Population Genetic Test Statistics
Pi
184.769254
Theta
167.921564
Tajima's D
-1.94458
CLR
139.803212
CSRT
0.0172991350432478
Interpretation
Possibly Positive selection
