Gene
KWMTBOMO08683
Pre Gene Modal
BGIBMGA007651
Annotation
PREDICTED:_prion-like-(Q/N-rich)_domain-bearing_protein_25_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.33
Sequence
CDS
ATGGGAGGATGGCAGCTGATTTTCGTGGCCCTCCTGGGCCTGGCGACAGCACAAGGCCCCGAGAAACGACGGACCGGTAACGTTTGCAACGTCGACATGGACTGTCCGGATAACGCTTTCTGCAGACTGAACAGTTACTGCGTCTGCAAGGAAGGGTACGTGTATGCAGCTATCAACAGCACTAACAAAGGATGCCTCAAAGAGGCCCGCAACGGGGAGGCGTGTGTCCAAAACATTCAGTGTCACATGACAATGGGTGTTCACTCTGAATGTACCGGAGGTAGCTGCGCGTGTGCTCCAACAGCTCATCTAGACGGGGAACGATGTTACGAGACTGCATTAATTGGTGAAAGATGTGTGGTCGATCAGAACTGCTACCTCGGCGAGCAAGGCATCGAAAGACAAGCATTCTGCGTACGAGGTTACTGTACATGTCAATTGCAGTTCAGCCCTAGGGACAATGGAACAAGATGCGTTAGAGACGCAGCTCTTGGCGAAGAATGCGAGGATCAGTTGCAATGCGCCGGACCTGGTTTGGAGTGCAGGGGTACTTGCAGATGCAGGGATAATTGGGTGGCCAACAACCACACCAACTCTTGCGTTGAATCCGTCAAAGCACTGAACGAACCGTGCGAATTCGACATACAGTGCGACTCGTTGGCCGGCGTTCCCGGCGTGCCGTCGCCCGGGGCAGCCCTGTGCCTCGGCGGGACCTGCTCGTGCTCTTACAACGCACGTCCGGTCGGGAACCCGCCGCGGTGCTGGCACAGGAGACGCCCGGGACAAGACTGCAGGAGAGATGAGGAATGCGTTTCTGAGGAAGACGAACCCGGTTATTGTTTGAACGGGCGGTGCACTTGCAAAACCTGTTCTCCTGACGCGCGGGACTTTAGTGGCGCGACGGTTTCCCGCCAATCGATGTTCGCGTCGGCCGTCGTGTTGTTCGCCGCCATTTTGGGATGA
Protein
MGGWQLIFVALLGLATAQGPEKRRTGNVCNVDMDCPDNAFCRLNSYCVCKEGYVYAAINSTNKGCLKEARNGEACVQNIQCHMTMGVHSECTGGSCACAPTAHLDGERCYETALIGERCVVDQNCYLGEQGIERQAFCVRGYCTCQLQFSPRDNGTRCVRDAALGEECEDQLQCAGPGLECRGTCRCRDNWVANNHTNSCVESVKALNEPCEFDIQCDSLAGVPGVPSPGAALCLGGTCSCSYNARPVGNPPRCWHRRRPGQDCRRDEECVSEEDEPGYCLNGRCTCKTCSPDARDFSGATVSRQSMFASAVVLFAAILG
Summary
Uniprot
A0A2A4JD57
A0A2W1BH98
A0A2H1VAH7
A0A194RAC6
A0A3S2LJF5
A0A1E1WIV8
+ More
A0A194PRN2 A0A212EGS9 A0A0L7LSB6 A0A2P8ZLP8 D6W9B3 A0A0M8ZSK9 A0A3L8DZ21 A0A1L8DNW8 A0A1L8DP39 A0A1L8DNV0 A0A1L8DNW6 A0A1L8DP60 A0A026WE43 A0A336M3Z1 A0A0L7RHE0 A0A336M7Z5 A0A182JD73 A0A182SJD4 A0A182YGF3 A0A182K7Q4 J3JZ96 R4FL12 A0A2M4CHL2 A0A2M4CH70 A0A023F6R2 A0A1W4WTN5 A0A1Y1LWA5 A0A182MG52 A0A182RNH6 A0A182N5C8 A0A1J1HYE6 A0A182UED5 A0A182HKR1 Q7PXG2 R4WCS4 A0A310STS3 A0A158NIH6 A0A182V1M6 A0A154PQG5 A0A182X9S9 Q17CJ7 A0A1Q3G3N7 N6UFC1 A0A182Q9A5 W5JIG1 A0A224XSW8 A0A0A9Z7K3 A0A1S4F8C0 A0A084WQU1 A0A182H0F3 A0A146M9P6 A0A0A9XKK9 A0A182VR84 A0A1B6MLP1 A0A1B6HEZ9 A0A1B6CDL3 A0A195BEM1 A0A1B6EII6 E0VUQ9 T1HWU2 R4FP86 R4UNK1 A0A224XSP3 A0A1S3JCL7 A0A1I8JU26 A0A224XNQ9
A0A194PRN2 A0A212EGS9 A0A0L7LSB6 A0A2P8ZLP8 D6W9B3 A0A0M8ZSK9 A0A3L8DZ21 A0A1L8DNW8 A0A1L8DP39 A0A1L8DNV0 A0A1L8DNW6 A0A1L8DP60 A0A026WE43 A0A336M3Z1 A0A0L7RHE0 A0A336M7Z5 A0A182JD73 A0A182SJD4 A0A182YGF3 A0A182K7Q4 J3JZ96 R4FL12 A0A2M4CHL2 A0A2M4CH70 A0A023F6R2 A0A1W4WTN5 A0A1Y1LWA5 A0A182MG52 A0A182RNH6 A0A182N5C8 A0A1J1HYE6 A0A182UED5 A0A182HKR1 Q7PXG2 R4WCS4 A0A310STS3 A0A158NIH6 A0A182V1M6 A0A154PQG5 A0A182X9S9 Q17CJ7 A0A1Q3G3N7 N6UFC1 A0A182Q9A5 W5JIG1 A0A224XSW8 A0A0A9Z7K3 A0A1S4F8C0 A0A084WQU1 A0A182H0F3 A0A146M9P6 A0A0A9XKK9 A0A182VR84 A0A1B6MLP1 A0A1B6HEZ9 A0A1B6CDL3 A0A195BEM1 A0A1B6EII6 E0VUQ9 T1HWU2 R4FP86 R4UNK1 A0A224XSP3 A0A1S3JCL7 A0A1I8JU26 A0A224XNQ9
Pubmed
EMBL
NWSH01001938
PCG69628.1
KZ150050
PZC74422.1
ODYU01001500
SOQ37796.1
+ More
KQ460473 KPJ14567.1 RSAL01000001 RVE55182.1 GDQN01004104 JAT86950.1 KQ459596 KPI95409.1 AGBW02015022 OWR40697.1 JTDY01000192 KOB78368.1 PYGN01000021 PSN57425.1 KQ971312 EEZ98472.1 KQ435876 KOX70117.1 QOIP01000002 RLU25365.1 GFDF01005938 JAV08146.1 GFDF01005937 JAV08147.1 GFDF01005931 JAV08153.1 GFDF01005932 JAV08152.1 GFDF01005930 JAV08154.1 KK107250 EZA54375.1 UFQS01000524 UFQT01000524 SSX04601.1 SSX24965.1 KQ414590 KOC70382.1 SSX04600.1 SSX24963.1 BT128577 AEE63534.1 GAHY01002284 JAA75226.1 GGFL01000561 MBW64739.1 GGFL01000323 MBW64501.1 GBBI01001661 JAC17051.1 GEZM01045351 JAV77812.1 AXCM01004258 CVRI01000026 CRK92356.1 APCN01000860 AAAB01008987 EAA01393.5 AK416959 BAN20174.1 KQ760478 OAD60220.1 ADTU01000264 KQ434999 KZC13350.1 CH477308 EAT44024.1 GFDL01000669 JAV34376.1 APGK01029195 APGK01029196 KB740727 KB631815 ENN79326.1 ERL86413.1 AXCN02001179 ADMH02001070 ETN64177.1 GFTR01005223 JAW11203.1 GBHO01005838 GBHO01005835 GBHO01005834 GBHO01005833 GBRD01011947 GDHC01015693 JAG37766.1 JAG37769.1 JAG37770.1 JAG37771.1 JAG53877.1 JAQ02936.1 ATLV01025730 ATLV01025731 ATLV01025732 ATLV01025733 ATLV01025734 ATLV01025735 ATLV01025736 ATLV01025737 ATLV01025738 KE525398 KFB52585.1 JXUM01101238 JXUM01101239 KQ564634 KXJ71982.1 GDHC01018434 GDHC01002138 JAQ00195.1 JAQ16491.1 GBHO01023100 GBHO01005839 JAG20504.1 JAG37765.1 GEBQ01003107 JAT36870.1 GECU01034460 JAS73246.1 GEDC01025761 JAS11537.1 KQ976509 KYM82659.1 GECZ01032032 JAS37737.1 DS235787 EEB17115.1 ACPB03005834 GAHY01000268 JAA77242.1 KC740926 AGM32750.1 GFTR01005435 JAW10991.1 GFTR01006633 JAW09793.1
KQ460473 KPJ14567.1 RSAL01000001 RVE55182.1 GDQN01004104 JAT86950.1 KQ459596 KPI95409.1 AGBW02015022 OWR40697.1 JTDY01000192 KOB78368.1 PYGN01000021 PSN57425.1 KQ971312 EEZ98472.1 KQ435876 KOX70117.1 QOIP01000002 RLU25365.1 GFDF01005938 JAV08146.1 GFDF01005937 JAV08147.1 GFDF01005931 JAV08153.1 GFDF01005932 JAV08152.1 GFDF01005930 JAV08154.1 KK107250 EZA54375.1 UFQS01000524 UFQT01000524 SSX04601.1 SSX24965.1 KQ414590 KOC70382.1 SSX04600.1 SSX24963.1 BT128577 AEE63534.1 GAHY01002284 JAA75226.1 GGFL01000561 MBW64739.1 GGFL01000323 MBW64501.1 GBBI01001661 JAC17051.1 GEZM01045351 JAV77812.1 AXCM01004258 CVRI01000026 CRK92356.1 APCN01000860 AAAB01008987 EAA01393.5 AK416959 BAN20174.1 KQ760478 OAD60220.1 ADTU01000264 KQ434999 KZC13350.1 CH477308 EAT44024.1 GFDL01000669 JAV34376.1 APGK01029195 APGK01029196 KB740727 KB631815 ENN79326.1 ERL86413.1 AXCN02001179 ADMH02001070 ETN64177.1 GFTR01005223 JAW11203.1 GBHO01005838 GBHO01005835 GBHO01005834 GBHO01005833 GBRD01011947 GDHC01015693 JAG37766.1 JAG37769.1 JAG37770.1 JAG37771.1 JAG53877.1 JAQ02936.1 ATLV01025730 ATLV01025731 ATLV01025732 ATLV01025733 ATLV01025734 ATLV01025735 ATLV01025736 ATLV01025737 ATLV01025738 KE525398 KFB52585.1 JXUM01101238 JXUM01101239 KQ564634 KXJ71982.1 GDHC01018434 GDHC01002138 JAQ00195.1 JAQ16491.1 GBHO01023100 GBHO01005839 JAG20504.1 JAG37765.1 GEBQ01003107 JAT36870.1 GECU01034460 JAS73246.1 GEDC01025761 JAS11537.1 KQ976509 KYM82659.1 GECZ01032032 JAS37737.1 DS235787 EEB17115.1 ACPB03005834 GAHY01000268 JAA77242.1 KC740926 AGM32750.1 GFTR01005435 JAW10991.1 GFTR01006633 JAW09793.1
Proteomes
UP000218220
UP000053240
UP000283053
UP000053268
UP000007151
UP000037510
+ More
UP000245037 UP000007266 UP000053105 UP000279307 UP000053097 UP000053825 UP000075880 UP000075901 UP000076408 UP000075881 UP000192223 UP000075883 UP000075900 UP000075884 UP000183832 UP000075902 UP000075840 UP000007062 UP000005205 UP000075903 UP000076502 UP000076407 UP000008820 UP000019118 UP000030742 UP000075886 UP000000673 UP000030765 UP000069940 UP000249989 UP000075920 UP000078540 UP000009046 UP000015103 UP000085678
UP000245037 UP000007266 UP000053105 UP000279307 UP000053097 UP000053825 UP000075880 UP000075901 UP000076408 UP000075881 UP000192223 UP000075883 UP000075900 UP000075884 UP000183832 UP000075902 UP000075840 UP000007062 UP000005205 UP000075903 UP000076502 UP000076407 UP000008820 UP000019118 UP000030742 UP000075886 UP000000673 UP000030765 UP000069940 UP000249989 UP000075920 UP000078540 UP000009046 UP000015103 UP000085678
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
A0A2A4JD57
A0A2W1BH98
A0A2H1VAH7
A0A194RAC6
A0A3S2LJF5
A0A1E1WIV8
+ More
A0A194PRN2 A0A212EGS9 A0A0L7LSB6 A0A2P8ZLP8 D6W9B3 A0A0M8ZSK9 A0A3L8DZ21 A0A1L8DNW8 A0A1L8DP39 A0A1L8DNV0 A0A1L8DNW6 A0A1L8DP60 A0A026WE43 A0A336M3Z1 A0A0L7RHE0 A0A336M7Z5 A0A182JD73 A0A182SJD4 A0A182YGF3 A0A182K7Q4 J3JZ96 R4FL12 A0A2M4CHL2 A0A2M4CH70 A0A023F6R2 A0A1W4WTN5 A0A1Y1LWA5 A0A182MG52 A0A182RNH6 A0A182N5C8 A0A1J1HYE6 A0A182UED5 A0A182HKR1 Q7PXG2 R4WCS4 A0A310STS3 A0A158NIH6 A0A182V1M6 A0A154PQG5 A0A182X9S9 Q17CJ7 A0A1Q3G3N7 N6UFC1 A0A182Q9A5 W5JIG1 A0A224XSW8 A0A0A9Z7K3 A0A1S4F8C0 A0A084WQU1 A0A182H0F3 A0A146M9P6 A0A0A9XKK9 A0A182VR84 A0A1B6MLP1 A0A1B6HEZ9 A0A1B6CDL3 A0A195BEM1 A0A1B6EII6 E0VUQ9 T1HWU2 R4FP86 R4UNK1 A0A224XSP3 A0A1S3JCL7 A0A1I8JU26 A0A224XNQ9
A0A194PRN2 A0A212EGS9 A0A0L7LSB6 A0A2P8ZLP8 D6W9B3 A0A0M8ZSK9 A0A3L8DZ21 A0A1L8DNW8 A0A1L8DP39 A0A1L8DNV0 A0A1L8DNW6 A0A1L8DP60 A0A026WE43 A0A336M3Z1 A0A0L7RHE0 A0A336M7Z5 A0A182JD73 A0A182SJD4 A0A182YGF3 A0A182K7Q4 J3JZ96 R4FL12 A0A2M4CHL2 A0A2M4CH70 A0A023F6R2 A0A1W4WTN5 A0A1Y1LWA5 A0A182MG52 A0A182RNH6 A0A182N5C8 A0A1J1HYE6 A0A182UED5 A0A182HKR1 Q7PXG2 R4WCS4 A0A310STS3 A0A158NIH6 A0A182V1M6 A0A154PQG5 A0A182X9S9 Q17CJ7 A0A1Q3G3N7 N6UFC1 A0A182Q9A5 W5JIG1 A0A224XSW8 A0A0A9Z7K3 A0A1S4F8C0 A0A084WQU1 A0A182H0F3 A0A146M9P6 A0A0A9XKK9 A0A182VR84 A0A1B6MLP1 A0A1B6HEZ9 A0A1B6CDL3 A0A195BEM1 A0A1B6EII6 E0VUQ9 T1HWU2 R4FP86 R4UNK1 A0A224XSP3 A0A1S3JCL7 A0A1I8JU26 A0A224XNQ9
Ontologies
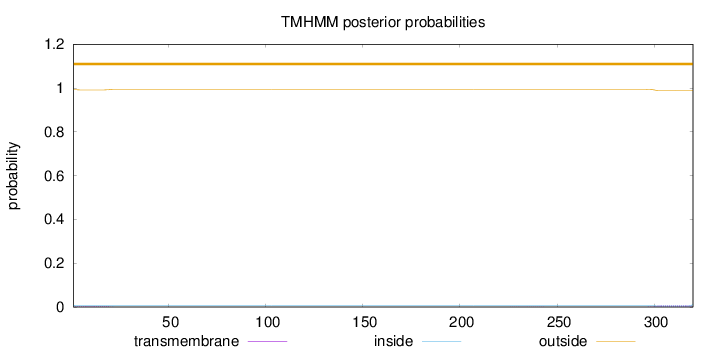
Topology
Subcellular location
Nucleus
SignalP
Position: 1 - 17,
Likelihood: 0.999522
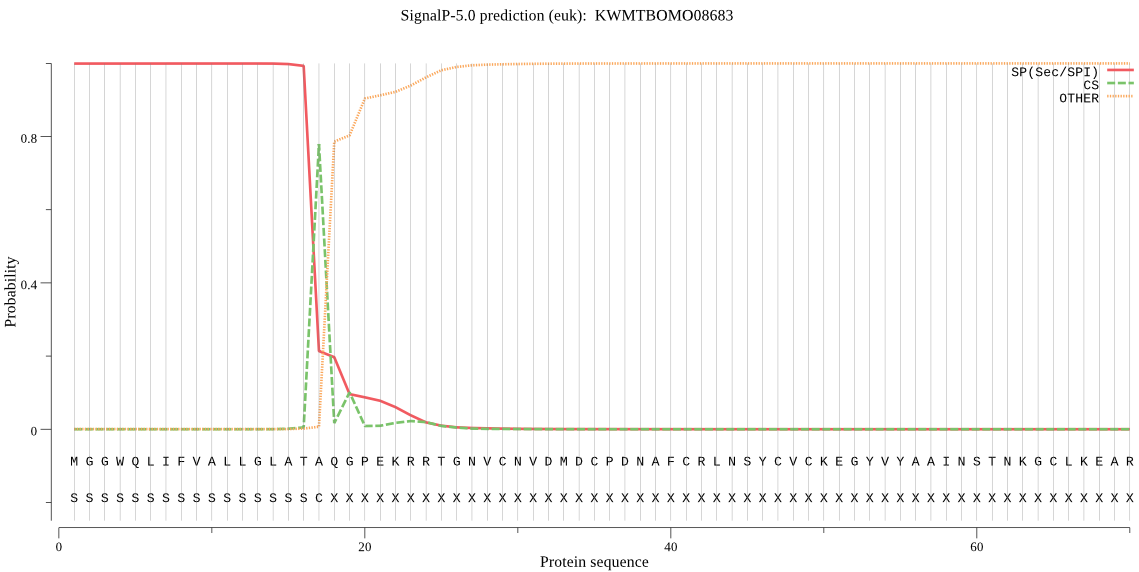
Length:
320
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21487
Exp number, first 60 AAs:
0.0608700000000001
Total prob of N-in:
0.00818
outside
1 - 320
Population Genetic Test Statistics
Pi
209.954675
Theta
170.575481
Tajima's D
0.383912
CLR
1.100574
CSRT
0.48177591120444
Interpretation
Uncertain
