Gene
KWMTBOMO08679 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007649
Annotation
PREDICTED:_uncharacterized_protein_LOC101742065_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.173
Sequence
CDS
ATGGCGTTCGTATTGTATATTTTGGCGGGCCTCCTGTTATCTGCGCACGCGCAATCGAACTACGGCTCCCAGGCCAACAACATCGAGTACCAAGGGGAAGGGCTGCCCGAACAAACTGTACTAGATGGAAAGGTCACGAAGCTGGACGACTTGAGTCCAGTTATCTTCTTGAACAGAACCAAAGCGGCACTGAACTGCGCTGCGGGTTCCATGCAGAACCCTCTGCGAGTATTCACGAACAACATAGTTGTAAGATTCCATCCTGGCTTAGAGATGGACGGAGACGAGGTCATCACAATCGTTTGTCGGTACCCACCGCCCATCGTTCCTAAGCCAGTCTTCAACCCGCTTATTGCCACTCCTGAACGCGGGCCGCTACCAGCGAAGGCTCCTTTAGCAGGAACGCATATCCTGATGATAATCTGCGCAATATTGTTTTTAACACTGCTGCTGCTTGGACTCGGAGTCTCATACTTGTGTCTGAGAAGACGCGCCTTGCCCGCTCCGAGGAGACTGATTGACGACTCCTCAGCGTCGATTGTGAGCAGAGAAAGTATACAAGAAGTGAAGATTCCTCGTGCACATCCTGTATACCCAATAGGCGGAGAGGCTGCGAGCGTGGCTTCGGATACCATTCCATCGGATTATCCATCAGAGACTCCCAGCGAGACCGACGCCGCTCATGCCAACCATGCTTTCATTATGGACGAAAACTATCACAGTGAAGGATATGGTGAAGCCCACGAGACTAGCGCTTCGAACGTGCTAGTGCACGCGGCGCCGCCCGCGTTCGACGTGCGCGTTCGCGTCCAGCGCCCCCCCGCGCCCCCCTCCCCGCAATCCACCATCACTACCACCGAGGTCGATGGTGGAAGCATGCGACACAGCAGCTACTTAGAGGAGCAAGCGCGGCAGGAGTCGGTCCGCACGGTGTCCCCGGTGGCGCTGCCGCTGCCCGCCCGCGCCGCGCAGCTGCCCCCGCCCGAGCGCCCCCCGCGCCCCCCCGCGCTCTATTCCCAGGTCAACAGAGAACGACAGGAGTGGTCGCGGCGGCCGCGGTCCATTGCCTCCCTCAACACGGAGATGACCGACACTCACTCGGTCACCGAAGTCACCGACAGGTCGCATGCTAGAATGTTCCATCTCAAGAAACCGCCCCCACCACCGCCGGTGATGAACGAACGTCTGGTGACGGAGGAACAGGAAGTAACGATGGAGAGCCTCGAGCCTCTTCCGGAGACACCCATCATGCCGGCCAGGAAGCCGGAGGTTACTTCACATGTCGTTGACGACGTGTTCCTGCGCACCATCACGGAAAAGAAGACCATCGAAGATGTCGAGCGACACAAGCGTCTCGTCACTGAATATAAACAGGTGCCGGCGCCGCCACCCCAGTTCGACGTGACGATCCGTAACCACACGGCCCCAGAGCCGCAGTGGGAGAACTTCTCGGACATAAGCAGCGCTTCCGGGATGACGCTGACGCCCAAGATGGAACGGGTACCGCTCTCGCTGCCCCCGCAGAAGATTATCGGCAAAGGGGGCCCGGACCTGTTCTCCCCGGAGTTGATCAAGCAGACGACTCATGTGTACCAGTCGCCGTCTTCTGCCGACCTCCCACTCCTGCCCGAGCTGCCTCCAGCCCGGAACCCTCTCTACAGGGACGAGGACGACGAGGATGTGCCGGAGCCGATTCCCGCACCACCAGTTCCAGCTAACTGGAGCGTTCTGACTAGAGTACTGAAAACTGAAGCTCAAGATGAGGTCCTGTTGGACTCGGAGCGCTCTCTGCGGCTGACGCGCGACGAGCGCCTCCGCTGGCGCGAGGTGATCACGCACGAGAGCACCCTGCGCCGCTCGTTGGCGCGTTCCTCCACCAGAGAGGACTTCACCAGAGTCGCCCACGACCAGCGCTACGCTCCCCTCTACACGCCTCACAAGTGGGACGTGATCATACGTATACTAGCGCCGCCCCACGACAAGCCGAAGAACCGCTACCGCAAGAAGACGGAGTGGGACTCCCGCTCCCGGCGTAGCTCGCTGCCGACGCTGTACGAGTATGACAGCGACGCCACGTCCCTGCGCGACCCGCAGCGCTCCCGCCGCTCCTCCTACCGAAGCGACCACGTTGACATGAGGTCGATGTCCGAAATGATGGTGGACTACGCGAGAGAGCAGGCGGACACGCACAGCGAAGTCTCCGGGGGAACTCAGCTCGGCAGGTACTACGATGACGACAGCGACTCCGAACATCCCTTCCATAATAACCAGTCGTGGTCCCGTCACTCTCTACACCGTTCAGCGAGTCAGCCGTCATTGGCGCGCTCCGCTACTGAAGTGGTGGAGCAGTGGACGGCGCCCGAGGGTGACGTCACACCCACACCGGGACGCAGGCTTAGAGGTCCTCAAGGCCTCACGACATTTACATCGTCTGAAGTACGAACGTTCCAAGCGACCTCCAGGGAATGGCGCGACTAA
Protein
MAFVLYILAGLLLSAHAQSNYGSQANNIEYQGEGLPEQTVLDGKVTKLDDLSPVIFLNRTKAALNCAAGSMQNPLRVFTNNIVVRFHPGLEMDGDEVITIVCRYPPPIVPKPVFNPLIATPERGPLPAKAPLAGTHILMIICAILFLTLLLLGLGVSYLCLRRRALPAPRRLIDDSSASIVSRESIQEVKIPRAHPVYPIGGEAASVASDTIPSDYPSETPSETDAAHANHAFIMDENYHSEGYGEAHETSASNVLVHAAPPAFDVRVRVQRPPAPPSPQSTITTTEVDGGSMRHSSYLEEQARQESVRTVSPVALPLPARAAQLPPPERPPRPPALYSQVNRERQEWSRRPRSIASLNTEMTDTHSVTEVTDRSHARMFHLKKPPPPPPVMNERLVTEEQEVTMESLEPLPETPIMPARKPEVTSHVVDDVFLRTITEKKTIEDVERHKRLVTEYKQVPAPPPQFDVTIRNHTAPEPQWENFSDISSASGMTLTPKMERVPLSLPPQKIIGKGGPDLFSPELIKQTTHVYQSPSSADLPLLPELPPARNPLYRDEDDEDVPEPIPAPPVPANWSVLTRVLKTEAQDEVLLDSERSLRLTRDERLRWREVITHESTLRRSLARSSTREDFTRVAHDQRYAPLYTPHKWDVIIRILAPPHDKPKNRYRKKTEWDSRSRRSSLPTLYEYDSDATSLRDPQRSRRSSYRSDHVDMRSMSEMMVDYAREQADTHSEVSGGTQLGRYYDDDSDSEHPFHNNQSWSRHSLHRSASQPSLARSATEVVEQWTAPEGDVTPTPGRRLRGPQGLTTFTSSEVRTFQATSREWRD
Summary
Uniprot
Pubmed
EMBL
BABH01003193
BABH01003194
KZ150060
PZC74240.1
ODYU01004761
SOQ44987.1
+ More
KQ460473 KPJ14571.1 NWSH01000177 PCG78715.1 RSAL01000001 RVE55184.1 PCG78714.1 AGBW02011461 OWR46612.1 KQ459596 KPI95405.1 JTDY01000990 KOB75184.1 LJIG01000453 KRT86487.1 GEZM01063240 JAV69168.1 APGK01041016 KB740987 KB631964 ENN76121.1 ERL87528.1 KQ971380 EEZ97267.1 GFDF01010856 JAV03228.1 CH899795 EAT32504.1 GFDL01002174 JAV32871.1 GECZ01001571 JAS68198.1 AXCN02001522 APCN01000876 AAAB01008987 EAA00847.5 GEDC01008097 JAS29201.1 AJVK01014014
KQ460473 KPJ14571.1 NWSH01000177 PCG78715.1 RSAL01000001 RVE55184.1 PCG78714.1 AGBW02011461 OWR46612.1 KQ459596 KPI95405.1 JTDY01000990 KOB75184.1 LJIG01000453 KRT86487.1 GEZM01063240 JAV69168.1 APGK01041016 KB740987 KB631964 ENN76121.1 ERL87528.1 KQ971380 EEZ97267.1 GFDF01010856 JAV03228.1 CH899795 EAT32504.1 GFDL01002174 JAV32871.1 GECZ01001571 JAS68198.1 AXCN02001522 APCN01000876 AAAB01008987 EAA00847.5 GEDC01008097 JAS29201.1 AJVK01014014
Proteomes
PRIDE
Pfam
PF00100 Zona_pellucida
Interpro
IPR001507
ZP_dom
ProteinModelPortal
Ontologies
GO
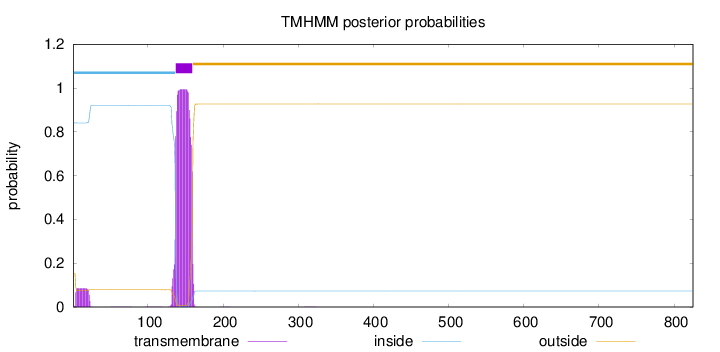
Topology
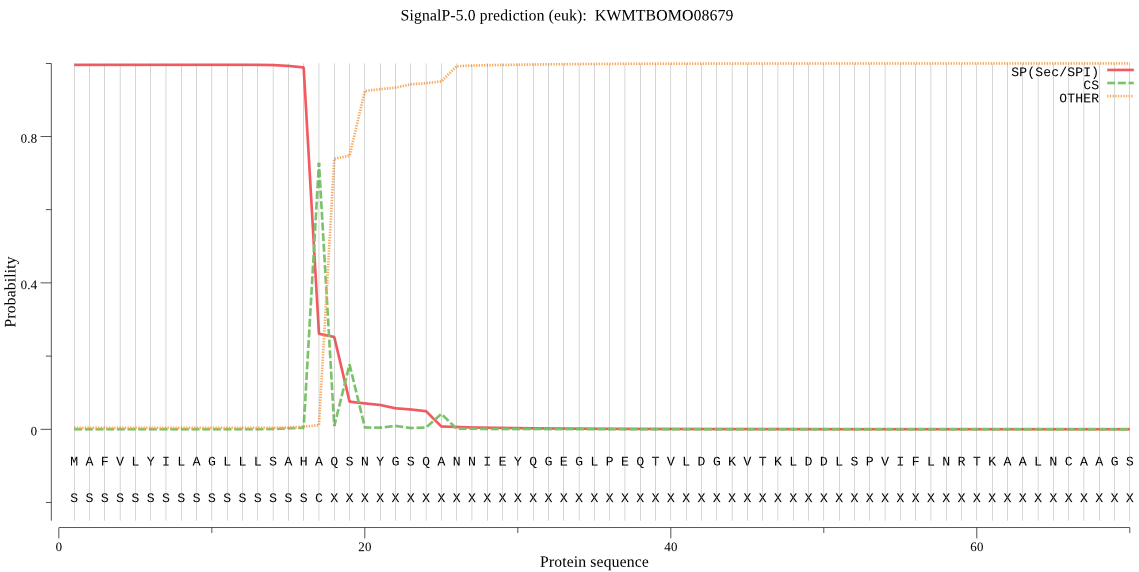
SignalP
Position: 1 - 17,
Likelihood: 0.995839
Length:
825
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.26099
Exp number, first 60 AAs:
1.60167
Total prob of N-in:
0.84566
inside
1 - 136
TMhelix
137 - 159
outside
160 - 825
Population Genetic Test Statistics
Pi
264.621529
Theta
162.294354
Tajima's D
1.802421
CLR
0.510429
CSRT
0.848707564621769
Interpretation
Uncertain
