Pre Gene Modal
BGIBMGA014236
Annotation
PREDICTED:_uncharacterized_protein_LOC105841512_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.101
Sequence
CDS
ATGCCGAATTCCGAGGAGCGCGACGCGAGAGCCGCGTCGCCGCCCCACGCGCCGTCGAGTGACGCGCCGCCCGCCAGTGTTTGCTTCCGCCCGGAAGACATCACCATGCTGATCGACACCCTTCAACGAACGCAGCTAGACGCGTTCCGGGAGCTGTTAGATAGCGTCAGGCAGTCGGCGACTCCTACATTCGACCATCGGCCCGCCAACAACTTCAGCCGCTGCAAATCTGTATTCAGCGGACGCGCAGACGAGTCAGTAGAAGGTTTTATCGACGCAATAGAATCTTACAGAGACGCCGCCAATGTTCCCGATGACGTAGCCGTAAAAGGCATCTCCATGTTGCTTACACACACCGCCGCTACCTGGTGGCAGGGTATCAAGCATCAAGTTTCTACATGGGAAGACGTCATCTCAAATCTACGAAGTGCTTTTGGTGATCGCCGCCCACCCCACAGAATTTATGTCGACCTATTCGCAGACGGCCAGCAGCCTATGGAGAAAACCGACCTGTTTGTCGCCAGGGCTCGAGCACTCCTCGCGAAGCTACCTCGAGGCGACTTATGTGAAAAAGTGGAGCTCGATATGATATACGGATTACTGCATCATAAAATTCGTAAACGCCTTCGTAGAGAGGAAATCACCAGTTTCAATGTTCTCCTGCAAAAAGCACGAAGTATTGAGGATTCTACGAAAGAAACGCATACTTCAAACGCCGTCGCCGAGACATCAAAGGGTGCCGCCGCCGCCGCCCGCGCTCGACTCGAGCCGCCGCGGCCTACAACGCAGCGTGTACCTACGGCCACTGTAGAAGTTTCCCCGCGCCAACGTTCGTCAGGCGATAACGCCGACGCCACTCCGACCGCCGAAAAACCCGAGAAACGTCGATACTGTGTTTATTGTAAAAGTTACGGTCATACGCGTGATCAGTGTCGTAAATTGTTATTAAAACAAACCGCGAATGACAGTAACGATAACGTACTCGCCGATGGTTCAATGCTAACACGACAAATGCTGACAACGACACTTGACGTGAAGTTATTACCGTGTAAAGTTATTCCCATGGAATTTGTCGTATTTCCGGACGCCCAGGATAATGACACATTATTGGGTGTAGACTTTTTAGTAGCAGCTGGTTTAGCCATCGACTTCCATACTTCGACATGGCGCTACCATGGTTCTGAACGAGTTTTTCCATTGGAGTTCGAGTACCCTAGTGACCTCACGCCTTGTTCTGCTGCTGATTTCTTAAGAGAAGACGAGGGAACTATGCTGTGTCCTCCAGAGCGTTAA
Protein
MPNSEERDARAASPPHAPSSDAPPASVCFRPEDITMLIDTLQRTQLDAFRELLDSVRQSATPTFDHRPANNFSRCKSVFSGRADESVEGFIDAIESYRDAANVPDDVAVKGISMLLTHTAATWWQGIKHQVSTWEDVISNLRSAFGDRRPPHRIYVDLFADGQQPMEKTDLFVARARALLAKLPRGDLCEKVELDMIYGLLHHKIRKRLRREEITSFNVLLQKARSIEDSTKETHTSNAVAETSKGAAAAARARLEPPRPTTQRVPTATVEVSPRQRSSGDNADATPTAEKPEKRRYCVYCKSYGHTRDQCRKLLLKQTANDSNDNVLADGSMLTRQMLTTTLDVKLLPCKVIPMEFVVFPDAQDNDTLLGVDFLVAAGLAIDFHTSTWRYHGSERVFPLEFEYPSDLTPCSAADFLREDEGTMLCPPER
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
EMBL
BABH01032771
BABH01032772
BABH01032773
BABH01032774
NWSH01003218
PCG66743.1
+ More
NWSH01003681 PCG65942.1 NWSH01000779 PCG74285.1 NWSH01002086 PCG69236.1 NWSH01000004 PCG81003.1 NWSH01000039 PCG80378.1 JTDY01010931 KOB57873.1 JTDY01004939 KOB67577.1 NWSH01000724 PCG74608.1 NWSH01004191 PCG65373.1 JTDY01006541 KOB65909.1 NWSH01002539 PCG68027.1 JXJN01013566 JTDY01007689 KOB65012.1 JTDY01006758 KOB65715.1
NWSH01003681 PCG65942.1 NWSH01000779 PCG74285.1 NWSH01002086 PCG69236.1 NWSH01000004 PCG81003.1 NWSH01000039 PCG80378.1 JTDY01010931 KOB57873.1 JTDY01004939 KOB67577.1 NWSH01000724 PCG74608.1 NWSH01004191 PCG65373.1 JTDY01006541 KOB65909.1 NWSH01002539 PCG68027.1 JXJN01013566 JTDY01007689 KOB65012.1 JTDY01006758 KOB65715.1
Proteomes
PRIDE
Interpro
IPR010926
Myosin_TH1
+ More
IPR036961 Kinesin_motor_dom_sf
IPR001609 Myosin_head_motor_dom
IPR027417 P-loop_NTPase
IPR021109 Peptidase_aspartic_dom_sf
IPR000048 IQ_motif_EF-hand-BS
IPR001969 Aspartic_peptidase_AS
IPR018061 Retropepsins
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR036961 Kinesin_motor_dom_sf
IPR001609 Myosin_head_motor_dom
IPR027417 P-loop_NTPase
IPR021109 Peptidase_aspartic_dom_sf
IPR000048 IQ_motif_EF-hand-BS
IPR001969 Aspartic_peptidase_AS
IPR018061 Retropepsins
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
Gene 3D
ProteinModelPortal
Ontologies
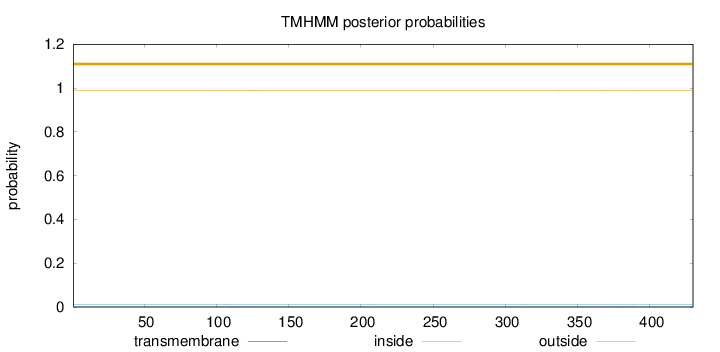
Topology
Length:
430
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02384
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01203
outside
1 - 430
Population Genetic Test Statistics
Pi
401.005776
Theta
177.41476
Tajima's D
3.916752
CLR
0.198926
CSRT
0.99830008499575
Interpretation
Uncertain
