Gene
KWMTBOMO08676
Annotation
PREDICTED:_RNA-directed_DNA_polymerase_from_mobile_element_jockey-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.414 PlasmaMembrane Reliability : 1.656
Sequence
CDS
ATGAATAGCAGCGGTTCTATTTATAATATAGTTAGTTTCAACTGCAAAGGAGTGAAACGATCCGTGGAACATGTAAGACAGCTGTGTCGGACGTCGGATATAATCGCGTTACAAGAGACGTGGTTATTGCCGGACGATTTACAGTTTCTCAGTGCCATTGATAGCGAATATGGCTGCACCGGGACTTCGGCCGTAGATACTGCGGCCGGGATACTCAGGGGGCGTCCATACGGAGGAGTGGCGCTACTATGGAAGAAAAGTATCTTTCAAAATGTGTCGATAGTGCAGTGTGATAACGCGCGTGTGTGTGCTATAAAAGTAACAATAGATCAGAGATCGTTATTGGTGTTCAGTATTTATATGCCTACGGATAATGCAGTGAACCTAACGGAGTTCATGGATTGCCTCAGCCTGATGAGCGCTATTATTAGTAACGAAAATATCGAATCAGTGTATATACTCGGCGATTTTAACGCCCATCCTAATGAACCTTTCTACAGGGAGTTGGCATGTGTCTGTGGGGAATTAGGATGGACTTGTGCGGACGTTGAACTATTAGGTGTAAATTCAGACACGTATAGTTTTATTAGTGAGGCGAATGGGTCCAGACGTTGGCTGGACCACTGTGTAGTCACGAAGTCAGCTATGCAGACCATTGTCAATACGTATATACTGTATGATGTCTTGTGGTCTGACCACTTTCCAGTAGTAGCACAGTGTGATTTTAACACTATAAAACCGAAATTTAATAATGTTACTCAATTGTGTAGTACTGTGCGATGGGGGGACAGAAAACCTGAGCAAATTGAATTTTATCATAAGATATGTCATGATAAACTGAGACAGATAGATTTTCCTGGCATTCTAAGTGATTGTTGTGATTGTGTTTGTAGTAATTCTGACCACAGACACATTTTAGATAAAATGTATCATGATATTGTTGACGTACTATGTCAAGCGGCTAGCGACAGTTATTCCCGTAGCAGATTTAATTGTAAAAAACCTGTTATTGGCTGGAATCGATATGTTAGTGATGCTCACGGGGAGGCGAGGTGTAAGTTTATTCTATGGGTGTGGCATGGTAAGCCGAGGTCGGGTCGTGTTTGGAAAGAAATGCAAGAGAGTAGGAAAATTTTTAAATCACGTCTGAAATGGTGTCAAAACCACAGCGAGCAGTTAAGAATGGATATACTCGCGTCTCATCACTCCAAGCACGATTTTCACAGCTTCTGGAAATCTACCACTAAACTTAATGGTAAACCAGGCCTGCCCGTGAGCGTCGGTGGTATAAGTGATCACGATGGCATCACGAATATGTTTAGGGAACACTTTGCGGTTAGTTCGCCATTGGGACCATCACAGTCAGGGTGTGTGGGTGATTCTCAGGGGCGGATAGTTACTGCTAGGTTTACGGCTAACGAAGTTAGAAAAGTTATTAACTCCATGTCACGGGGTAAGTCTCCGGGTCATGACGAGCTGAGCATAGAACACCTAAAGTATGCCGGTGTACACTTGCCCCGAGTCTTGGTTATGTTCTACAATTTATGTATAGGACACTCCTACTTGCCTGAGGAGATGTTGAAGACTGTGGTAGTGCCAATTGTGAAGAATAGGACCGGGGATATATGTGATAAAACCAATTACAGACCCATCTCGTTAGCAACTATCATAGCGAAAGTGCTTGACAGTTTGCTTAATATACAACTTGATAAATATCTTCAGATACATGATAACCAGTTTGGTTTTAAATCAGGAATGTCTACAGAAATAGCCGTATTGTGTTTAAAGCAAACTGTCAAGTACTATACTGATCGAAAAACTCCAATTTTTGCCTGTTTCCTCGATTTATCAAAGGCATTCGACCTGGTTTGTTATGACATCTTATGGCAAAAACTGAATAATACTAAAATGCCGAAAGAACTAGTACAGATATTCAAATATTGGTACGCCAACCAGCTGAACGTGGTACGATGGTCGGGTGTCTATTCGAGGCCATACGGGATGGAGTGCGGGGTGAGGCAAGGGGGGTTGACCTCGCCCAAGCTCTTCAACCTGTATGTGAATGCTCTAATCGAGGAGCTCAGCAGTACTCATGTCGGCTGCCATATTGATGAAATATGTCTCAACAATATCAGTTATGCTGATGACATGGTGCTGCTGAGTGCGTCGGCCTGTGGGCTGTCTAAGCTGCTTGGCATATGTGAAAGGTATGCTCTAAGTCACGGTCTGGTGTATAATACAAAGAAAAGTGAGTGTATGGTGTTTCAGGCCAGAGGCAGGTGTCTGGATAACATTCCCTCCGTCAAACTGAATGGTACGCCACTGCGCAGAGTAGACAAGTTTAAATACCTAGGACACTTTGTAACCTCTGACCTGAGGGATGATACAGACATTGAGCGGGAGCGGAGAGCGCTGTCTCCTGTGGGTGTTCTATAG
Protein
MNSSGSIYNIVSFNCKGVKRSVEHVRQLCRTSDIIALQETWLLPDDLQFLSAIDSEYGCTGTSAVDTAAGILRGRPYGGVALLWKKSIFQNVSIVQCDNARVCAIKVTIDQRSLLVFSIYMPTDNAVNLTEFMDCLSLMSAIISNENIESVYILGDFNAHPNEPFYRELACVCGELGWTCADVELLGVNSDTYSFISEANGSRRWLDHCVVTKSAMQTIVNTYILYDVLWSDHFPVVAQCDFNTIKPKFNNVTQLCSTVRWGDRKPEQIEFYHKICHDKLRQIDFPGILSDCCDCVCSNSDHRHILDKMYHDIVDVLCQAASDSYSRSRFNCKKPVIGWNRYVSDAHGEARCKFILWVWHGKPRSGRVWKEMQESRKIFKSRLKWCQNHSEQLRMDILASHHSKHDFHSFWKSTTKLNGKPGLPVSVGGISDHDGITNMFREHFAVSSPLGPSQSGCVGDSQGRIVTARFTANEVRKVINSMSRGKSPGHDELSIEHLKYAGVHLPRVLVMFYNLCIGHSYLPEEMLKTVVVPIVKNRTGDICDKTNYRPISLATIIAKVLDSLLNIQLDKYLQIHDNQFGFKSGMSTEIAVLCLKQTVKYYTDRKTPIFACFLDLSKAFDLVCYDILWQKLNNTKMPKELVQIFKYWYANQLNVVRWSGVYSRPYGMECGVRQGGLTSPKLFNLYVNALIEELSSTHVGCHIDEICLNNISYADDMVLLSASACGLSKLLGICERYALSHGLVYNTKKSECMVFQARGRCLDNIPSVKLNGTPLRRVDKFKYLGHFVTSDLRDDTDIERERRALSPVGVL
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
RSAL01000132
RVE46368.1
KZ149956
PZC76465.1
KZ150039
PZC74591.1
+ More
GAIX01014011 JAA78549.1 KZ150625 PZC70440.1 HAEJ01003895 SBS44352.1 GCES01018090 JAR68233.1 GBYX01473474 GBYX01473473 GBYX01473472 GBYX01473468 GBYX01473467 GBYX01473465 JAO08192.1 GDRN01099830 JAI58698.1 DMQF01000358 HAO15222.1 DMQF01000507 HAO15811.1 DMQF01000051 HAO14134.1 DNUT01000105 HBI39711.1 DNUT01000366 HBI40674.1 GEZM01098732 JAV53782.1 GDHC01021277 JAP97351.1 GBHO01042545 GBRD01013465 JAG01059.1 JAG52361.1 REGN01005167 RNA14504.1 GFTR01008507 JAW07919.1
GAIX01014011 JAA78549.1 KZ150625 PZC70440.1 HAEJ01003895 SBS44352.1 GCES01018090 JAR68233.1 GBYX01473474 GBYX01473473 GBYX01473472 GBYX01473468 GBYX01473467 GBYX01473465 JAO08192.1 GDRN01099830 JAI58698.1 DMQF01000358 HAO15222.1 DMQF01000507 HAO15811.1 DMQF01000051 HAO14134.1 DNUT01000105 HBI39711.1 DNUT01000366 HBI40674.1 GEZM01098732 JAV53782.1 GDHC01021277 JAP97351.1 GBHO01042545 GBRD01013465 JAG01059.1 JAG52361.1 REGN01005167 RNA14504.1 GFTR01008507 JAW07919.1
Proteomes
Pfam
Interpro
IPR024989
MFS_assoc_dom
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR036259 MFS_trans_sf
IPR000477 RT_dom
IPR020846 MFS_dom
IPR033116 TRYPSIN_SER
IPR036790 Frizzled_dom_sf
IPR020067 Frizzled_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR001190 SRCR
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR036259 MFS_trans_sf
IPR000477 RT_dom
IPR020846 MFS_dom
IPR033116 TRYPSIN_SER
IPR036790 Frizzled_dom_sf
IPR020067 Frizzled_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR001190 SRCR
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
SUPFAM
Gene 3D
ProteinModelPortal
PDB
5HHL
E-value=0.00102315,
Score=103
Ontologies
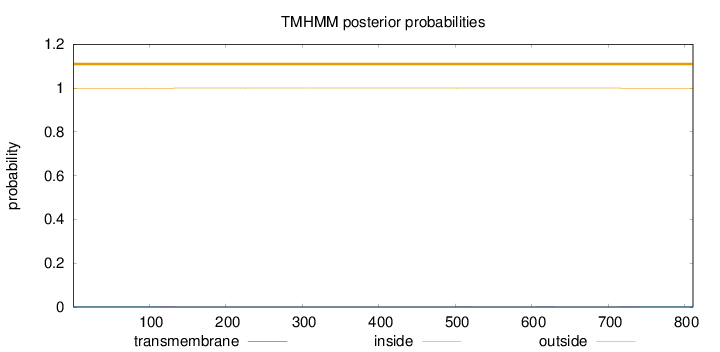
Topology
Length:
811
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03178
Exp number, first 60 AAs:
3e-05
Total prob of N-in:
0.00062
outside
1 - 811
Population Genetic Test Statistics
Pi
37.160711
Theta
49.292578
Tajima's D
-0.716282
CLR
242.308164
CSRT
0.198890055497225
Interpretation
Uncertain
