Gene
KWMTBOMO08674
Pre Gene Modal
BGIBMGA007934
Annotation
PREDICTED:_probable_Xaa-Pro_aminopeptidase_3_isoform_X1_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 2.399
Sequence
CDS
ATGCAGCGACTACGTAAAGTACTTCCATCTTTGGCCGTAAAGCATCACAAACTACTTAAGTCTCATATAAGAAATCAACTTAGATGCCTTTCTACAATTGACCACCCCCCTGGAGCGGTAGAGAGACCTACACTCTCTATACCACAAGGCACCTTCGGCCAAGCAACCTGTCACACCCATCCGCACTTGATCCAACAGGGGCATCTTACTTGCGGGATCACACAAAAAGAATATGCGGAGAGGAGAGAAACCTTAATTAGCAGACTTGTGAGTGAAGCTCCGAATGTGCACAAAACACATATTGTAGTAATCCCAGCTGCCCGCAAACAATTCATGTCTGATAAAATACCTTATGTGTTTAGACAGAATTCGGACTTTTTCTATCTTACAGGATGTTTAGAACCTTCAGCTATACTAGTCATGATTAAGCCAGCACACAGTGATACTTTTAAGAGTGTACTATTTGTTCACGAGAAAGACAGTCATGCAGAACTGTGGGAAGGTCCTCGAACTGGCTGCACGGCTGCCACACAGCTGTTTGCTGTGGACGAAGCTAGACCTGTCGACAGCTTTAGTGTATACATCAATAAGTTAGCGGCATCATCCAAACCTTCTGTCCTGTGGTACCACAACGAAGCACCTGCAAATCCTGAAATCCACAGCACGATACGCTCCGTGATACATCCTGAAACACAGGTGACGTTGGCGGATCCACAAAGAACGCTTCACTTCATGAGGGTGATCAAGTCTCCAGCCGAAGTGGAGCTGATGAAGGAGGTGTGCTACATTGGGTCACAGAGCGTTAACACAGCAATGGCCTGCACCAAACCCGGTATATCGGAGCACGCGGTGTGCTCGGTGCTGGAGCACAGCGGGCGCGTGAGTGGCGCGGAGCACAGTTCGTTCCCGGCCGTGGTCGCGGGGGCCGCCCGGGCCGCGCACATACACTACGTCGCCAACAACCAGCTGCTCAACCACGGGGACCTGCTGCTCGTCGACTCTGGCTGTCAGCGTTGGTTGTACAACTCTGACATCTCCCGCACGTGGCCGGTCTCCGGACTCTTCACGCGCCACCAGAGGATCCTGTACGAGCTGGTGCTCGCCGTGCAGAAACGCTTAATAGAAATCCTGGGGGAGCACCGCCCAGCGCTGGACCAGCTGTTCGACAGCATGTGCCGGCTGCTGGGCACGCAGCTGCAGCAAGAGGGGATACTGCCCAAGAACATTGATGGGCACGAACTGATCTCGATGGCGTACAGCCTGTGTCCGCACCACGTGTCGCACCACCTGGGGCTGGACGTGCACGACGCGCCGCTGGTCCGCCGCGCCACGCCCGTGCTCGCCAACATGATCGTCACTGTCGAGCCCGGTATCTACATCCGGACGGACGACACCCGGGTGCCGGAGGAGTTCCGCGGCCTGGGTATCCGTATAGAAGACGACGTCCTCATCACGGACGGTGACCCGCTGGTGCTCACCGACTCCTGCGCCAAGGAGGTGCAGGATATAGAAGCCATAGTCGGGAAACAGGCTTACTGA
Protein
MQRLRKVLPSLAVKHHKLLKSHIRNQLRCLSTIDHPPGAVERPTLSIPQGTFGQATCHTHPHLIQQGHLTCGITQKEYAERRETLISRLVSEAPNVHKTHIVVIPAARKQFMSDKIPYVFRQNSDFFYLTGCLEPSAILVMIKPAHSDTFKSVLFVHEKDSHAELWEGPRTGCTAATQLFAVDEARPVDSFSVYINKLAASSKPSVLWYHNEAPANPEIHSTIRSVIHPETQVTLADPQRTLHFMRVIKSPAEVELMKEVCYIGSQSVNTAMACTKPGISEHAVCSVLEHSGRVSGAEHSSFPAVVAGAARAAHIHYVANNQLLNHGDLLLVDSGCQRWLYNSDISRTWPVSGLFTRHQRILYELVLAVQKRLIEILGEHRPALDQLFDSMCRLLGTQLQQEGILPKNIDGHELISMAYSLCPHHVSHHLGLDVHDAPLVRRATPVLANMIVTVEPGIYIRTDDTRVPEEFRGLGIRIEDDVLITDGDPLVLTDSCAKEVQDIEAIVGKQAY
Summary
Cofactor
Mn(2+)
Similarity
Belongs to the peptidase M24B family.
Uniprot
A0A2H1W9B4
A0A0N1IIK9
A0A2A4K371
A0A194PR17
A0A2A4K4X0
A0A3S2P8T2
+ More
A0A1W4X3V9 K7IQ45 A0A232FAU8 A0A0T6AZQ9 E9IMB0 A0A195EMT4 A0A195CQG5 A0A151X5Y8 A0A026WL28 D6X4Q9 A0A195F946 A0A158NCL0 E2BM74 A0A1Y1KRI0 A0A195BXH1 A0A2J7PC92 D3TND4 A0A1A9XKG6 A0A0L7R510 A0A1B0AFD8 A0A1A9VNE7 A0A1B0AUC2 A0A1I8Q9X7 A0A088AL12 A0A0L0C8K8 W8B136 A0A1A9WA68 A0A067QY78 A0A1B6CM13 A0A0A9W1T7 A0A1B6MA28 A0A1S4FRT9 Q17F24 Q16RG9 A0A182GWY8 A0A1B0GKB0 A0A1I8MY31 A0A182UQJ0 A0A310SK25 A0A182WY26 A0A182TRP3 B4NE82 A0A182KW95 Q7Q4S9 A0A1I8QA11 A0A182HMT7 A0A084WPV6 A0A1Q3FA31 A0A0K8VC54 A0A0N0BBU3 A0A1Q1M8J6 A0A2A3E1R4 A0A182JGR5 A0A1B6I9T9 A0A1B6ELJ3 A0A1B0DC12 B0WPD2 A0A1S5QN16 A0A182N1L3 A0A336LME4 B3MQI7 A0A1J1HTU4 A0A336M323 N6UTE2 A0A182Q3P5 E0VMS9 A0A182RM20 A0A154P7L6 A0A293M5D4 B4L7X5 A0A182VV45 J9K7L6 A0A1Q3FA72 A0A2R5LEJ9 E2AK79 A0A1W4US83 A0A2M4CVR1 A0A1Q1M8J9 A0A2M4CVW1 Q29GW0 A0A182FIJ2 A0A3B0JXK4 A0A224YFX9 A0A2H8TQ15 W4YTF9 A0A1Q1M8M9 A0A131YXS3 A0A131XE35 A0A1Q1M8K1 L7M1Z9 Q9W5W7 B4M9Y0
A0A1W4X3V9 K7IQ45 A0A232FAU8 A0A0T6AZQ9 E9IMB0 A0A195EMT4 A0A195CQG5 A0A151X5Y8 A0A026WL28 D6X4Q9 A0A195F946 A0A158NCL0 E2BM74 A0A1Y1KRI0 A0A195BXH1 A0A2J7PC92 D3TND4 A0A1A9XKG6 A0A0L7R510 A0A1B0AFD8 A0A1A9VNE7 A0A1B0AUC2 A0A1I8Q9X7 A0A088AL12 A0A0L0C8K8 W8B136 A0A1A9WA68 A0A067QY78 A0A1B6CM13 A0A0A9W1T7 A0A1B6MA28 A0A1S4FRT9 Q17F24 Q16RG9 A0A182GWY8 A0A1B0GKB0 A0A1I8MY31 A0A182UQJ0 A0A310SK25 A0A182WY26 A0A182TRP3 B4NE82 A0A182KW95 Q7Q4S9 A0A1I8QA11 A0A182HMT7 A0A084WPV6 A0A1Q3FA31 A0A0K8VC54 A0A0N0BBU3 A0A1Q1M8J6 A0A2A3E1R4 A0A182JGR5 A0A1B6I9T9 A0A1B6ELJ3 A0A1B0DC12 B0WPD2 A0A1S5QN16 A0A182N1L3 A0A336LME4 B3MQI7 A0A1J1HTU4 A0A336M323 N6UTE2 A0A182Q3P5 E0VMS9 A0A182RM20 A0A154P7L6 A0A293M5D4 B4L7X5 A0A182VV45 J9K7L6 A0A1Q3FA72 A0A2R5LEJ9 E2AK79 A0A1W4US83 A0A2M4CVR1 A0A1Q1M8J9 A0A2M4CVW1 Q29GW0 A0A182FIJ2 A0A3B0JXK4 A0A224YFX9 A0A2H8TQ15 W4YTF9 A0A1Q1M8M9 A0A131YXS3 A0A131XE35 A0A1Q1M8K1 L7M1Z9 Q9W5W7 B4M9Y0
Pubmed
26354079
20075255
28648823
21282665
24508170
30249741
+ More
18362917 19820115 21347285 20798317 28004739 20353571 26108605 24495485 24845553 25401762 17510324 26483478 25315136 17994087 20966253 12364791 14747013 17210077 24438588 28126515 23537049 20566863 15632085 28797301 26830274 28049606 25576852 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
18362917 19820115 21347285 20798317 28004739 20353571 26108605 24495485 24845553 25401762 17510324 26483478 25315136 17994087 20966253 12364791 14747013 17210077 24438588 28126515 23537049 20566863 15632085 28797301 26830274 28049606 25576852 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
ODYU01007147
SOQ49671.1
KQ459781
KPJ19963.1
NWSH01000177
PCG78711.1
+ More
KQ459596 KPI95403.1 PCG78712.1 RSAL01000001 RVE55186.1 NNAY01000571 OXU27610.1 LJIG01016437 KRT80604.1 GL764129 EFZ18290.1 KQ978691 KYN29212.1 KQ977408 KYN02951.1 KQ982490 KYQ55796.1 KK107167 QOIP01000010 EZA56376.1 RLU17869.1 KQ971380 EEZ97679.2 KQ981727 KYN36963.1 ADTU01011955 GL449158 EFN83200.1 GEZM01075720 JAV64003.1 KQ976396 KYM92995.1 NEVH01027067 PNF13953.1 CCAG010006700 EZ422936 ADD19212.1 KQ414654 KOC65914.1 JXJN01003606 JRES01000757 KNC28611.1 GAMC01015822 JAB90733.1 KK853205 KDR09847.1 GEDC01022781 JAS14517.1 GBHO01041207 GBRD01002466 JAG02397.1 JAG63355.1 GEBQ01007210 JAT32767.1 CH477277 EAT45107.1 CH477710 EAT37000.1 JXUM01094253 KQ564116 KXJ72749.1 AJWK01028644 KQ761783 OAD57036.1 CH964239 EDW82051.2 AAAB01008963 EAA12035.4 APCN01004904 ATLV01025133 KE525369 KFB52250.1 GFDL01010673 JAV24372.1 GDHF01031058 GDHF01016204 JAI21256.1 JAI36110.1 KQ435989 KOX67691.1 KY295718 AQM32413.1 KZ288445 PBC25640.1 GECU01024029 JAS83677.1 GECZ01030972 JAS38797.1 AJVK01030483 DS232022 EDS32307.1 KR068501 AMO02518.1 UFQT01000059 SSX19156.1 CH902621 EDV44613.1 CVRI01000013 CRK89593.1 UFQS01000478 UFQT01000478 SSX04280.1 SSX24645.1 APGK01017071 KB739995 ENN82032.1 AXCN02002449 DS235324 EEB14685.1 KQ434831 KZC07833.1 GFWV01010529 MAA35258.1 CH933814 EDW05550.2 ABLF02024823 GFDL01010560 JAV24485.1 GGLE01003752 MBY07878.1 GL440194 EFN66183.1 GGFL01005232 MBW69410.1 KY295724 AQM32419.1 GGFL01005231 MBW69409.1 CH379064 EAL31999.2 OUUW01000003 SPP78499.1 GFPF01005452 MAA16598.1 GFXV01003533 MBW15338.1 AAGJ04051368 KY295723 AQM32418.1 GEDV01004558 JAP83999.1 GEFH01004201 JAP64380.1 KY295725 AQM32420.1 GACK01007886 JAA57148.1 AE014298 AY069164 AAF45368.1 AAL39309.1 CH940655 EDW66039.1
KQ459596 KPI95403.1 PCG78712.1 RSAL01000001 RVE55186.1 NNAY01000571 OXU27610.1 LJIG01016437 KRT80604.1 GL764129 EFZ18290.1 KQ978691 KYN29212.1 KQ977408 KYN02951.1 KQ982490 KYQ55796.1 KK107167 QOIP01000010 EZA56376.1 RLU17869.1 KQ971380 EEZ97679.2 KQ981727 KYN36963.1 ADTU01011955 GL449158 EFN83200.1 GEZM01075720 JAV64003.1 KQ976396 KYM92995.1 NEVH01027067 PNF13953.1 CCAG010006700 EZ422936 ADD19212.1 KQ414654 KOC65914.1 JXJN01003606 JRES01000757 KNC28611.1 GAMC01015822 JAB90733.1 KK853205 KDR09847.1 GEDC01022781 JAS14517.1 GBHO01041207 GBRD01002466 JAG02397.1 JAG63355.1 GEBQ01007210 JAT32767.1 CH477277 EAT45107.1 CH477710 EAT37000.1 JXUM01094253 KQ564116 KXJ72749.1 AJWK01028644 KQ761783 OAD57036.1 CH964239 EDW82051.2 AAAB01008963 EAA12035.4 APCN01004904 ATLV01025133 KE525369 KFB52250.1 GFDL01010673 JAV24372.1 GDHF01031058 GDHF01016204 JAI21256.1 JAI36110.1 KQ435989 KOX67691.1 KY295718 AQM32413.1 KZ288445 PBC25640.1 GECU01024029 JAS83677.1 GECZ01030972 JAS38797.1 AJVK01030483 DS232022 EDS32307.1 KR068501 AMO02518.1 UFQT01000059 SSX19156.1 CH902621 EDV44613.1 CVRI01000013 CRK89593.1 UFQS01000478 UFQT01000478 SSX04280.1 SSX24645.1 APGK01017071 KB739995 ENN82032.1 AXCN02002449 DS235324 EEB14685.1 KQ434831 KZC07833.1 GFWV01010529 MAA35258.1 CH933814 EDW05550.2 ABLF02024823 GFDL01010560 JAV24485.1 GGLE01003752 MBY07878.1 GL440194 EFN66183.1 GGFL01005232 MBW69410.1 KY295724 AQM32419.1 GGFL01005231 MBW69409.1 CH379064 EAL31999.2 OUUW01000003 SPP78499.1 GFPF01005452 MAA16598.1 GFXV01003533 MBW15338.1 AAGJ04051368 KY295723 AQM32418.1 GEDV01004558 JAP83999.1 GEFH01004201 JAP64380.1 KY295725 AQM32420.1 GACK01007886 JAA57148.1 AE014298 AY069164 AAF45368.1 AAL39309.1 CH940655 EDW66039.1
Proteomes
UP000053240
UP000218220
UP000053268
UP000283053
UP000192223
UP000002358
+ More
UP000215335 UP000078492 UP000078542 UP000075809 UP000053097 UP000279307 UP000007266 UP000078541 UP000005205 UP000008237 UP000078540 UP000235965 UP000092444 UP000092443 UP000053825 UP000092445 UP000078200 UP000092460 UP000095300 UP000005203 UP000037069 UP000091820 UP000027135 UP000008820 UP000069940 UP000249989 UP000092461 UP000095301 UP000075903 UP000076407 UP000075902 UP000007798 UP000075882 UP000007062 UP000075840 UP000030765 UP000053105 UP000242457 UP000075880 UP000092462 UP000002320 UP000075884 UP000007801 UP000183832 UP000019118 UP000075886 UP000009046 UP000075900 UP000076502 UP000009192 UP000075920 UP000007819 UP000000311 UP000192221 UP000001819 UP000069272 UP000268350 UP000007110 UP000000803 UP000008792
UP000215335 UP000078492 UP000078542 UP000075809 UP000053097 UP000279307 UP000007266 UP000078541 UP000005205 UP000008237 UP000078540 UP000235965 UP000092444 UP000092443 UP000053825 UP000092445 UP000078200 UP000092460 UP000095300 UP000005203 UP000037069 UP000091820 UP000027135 UP000008820 UP000069940 UP000249989 UP000092461 UP000095301 UP000075903 UP000076407 UP000075902 UP000007798 UP000075882 UP000007062 UP000075840 UP000030765 UP000053105 UP000242457 UP000075880 UP000092462 UP000002320 UP000075884 UP000007801 UP000183832 UP000019118 UP000075886 UP000009046 UP000075900 UP000076502 UP000009192 UP000075920 UP000007819 UP000000311 UP000192221 UP000001819 UP000069272 UP000268350 UP000007110 UP000000803 UP000008792
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1W9B4
A0A0N1IIK9
A0A2A4K371
A0A194PR17
A0A2A4K4X0
A0A3S2P8T2
+ More
A0A1W4X3V9 K7IQ45 A0A232FAU8 A0A0T6AZQ9 E9IMB0 A0A195EMT4 A0A195CQG5 A0A151X5Y8 A0A026WL28 D6X4Q9 A0A195F946 A0A158NCL0 E2BM74 A0A1Y1KRI0 A0A195BXH1 A0A2J7PC92 D3TND4 A0A1A9XKG6 A0A0L7R510 A0A1B0AFD8 A0A1A9VNE7 A0A1B0AUC2 A0A1I8Q9X7 A0A088AL12 A0A0L0C8K8 W8B136 A0A1A9WA68 A0A067QY78 A0A1B6CM13 A0A0A9W1T7 A0A1B6MA28 A0A1S4FRT9 Q17F24 Q16RG9 A0A182GWY8 A0A1B0GKB0 A0A1I8MY31 A0A182UQJ0 A0A310SK25 A0A182WY26 A0A182TRP3 B4NE82 A0A182KW95 Q7Q4S9 A0A1I8QA11 A0A182HMT7 A0A084WPV6 A0A1Q3FA31 A0A0K8VC54 A0A0N0BBU3 A0A1Q1M8J6 A0A2A3E1R4 A0A182JGR5 A0A1B6I9T9 A0A1B6ELJ3 A0A1B0DC12 B0WPD2 A0A1S5QN16 A0A182N1L3 A0A336LME4 B3MQI7 A0A1J1HTU4 A0A336M323 N6UTE2 A0A182Q3P5 E0VMS9 A0A182RM20 A0A154P7L6 A0A293M5D4 B4L7X5 A0A182VV45 J9K7L6 A0A1Q3FA72 A0A2R5LEJ9 E2AK79 A0A1W4US83 A0A2M4CVR1 A0A1Q1M8J9 A0A2M4CVW1 Q29GW0 A0A182FIJ2 A0A3B0JXK4 A0A224YFX9 A0A2H8TQ15 W4YTF9 A0A1Q1M8M9 A0A131YXS3 A0A131XE35 A0A1Q1M8K1 L7M1Z9 Q9W5W7 B4M9Y0
A0A1W4X3V9 K7IQ45 A0A232FAU8 A0A0T6AZQ9 E9IMB0 A0A195EMT4 A0A195CQG5 A0A151X5Y8 A0A026WL28 D6X4Q9 A0A195F946 A0A158NCL0 E2BM74 A0A1Y1KRI0 A0A195BXH1 A0A2J7PC92 D3TND4 A0A1A9XKG6 A0A0L7R510 A0A1B0AFD8 A0A1A9VNE7 A0A1B0AUC2 A0A1I8Q9X7 A0A088AL12 A0A0L0C8K8 W8B136 A0A1A9WA68 A0A067QY78 A0A1B6CM13 A0A0A9W1T7 A0A1B6MA28 A0A1S4FRT9 Q17F24 Q16RG9 A0A182GWY8 A0A1B0GKB0 A0A1I8MY31 A0A182UQJ0 A0A310SK25 A0A182WY26 A0A182TRP3 B4NE82 A0A182KW95 Q7Q4S9 A0A1I8QA11 A0A182HMT7 A0A084WPV6 A0A1Q3FA31 A0A0K8VC54 A0A0N0BBU3 A0A1Q1M8J6 A0A2A3E1R4 A0A182JGR5 A0A1B6I9T9 A0A1B6ELJ3 A0A1B0DC12 B0WPD2 A0A1S5QN16 A0A182N1L3 A0A336LME4 B3MQI7 A0A1J1HTU4 A0A336M323 N6UTE2 A0A182Q3P5 E0VMS9 A0A182RM20 A0A154P7L6 A0A293M5D4 B4L7X5 A0A182VV45 J9K7L6 A0A1Q3FA72 A0A2R5LEJ9 E2AK79 A0A1W4US83 A0A2M4CVR1 A0A1Q1M8J9 A0A2M4CVW1 Q29GW0 A0A182FIJ2 A0A3B0JXK4 A0A224YFX9 A0A2H8TQ15 W4YTF9 A0A1Q1M8M9 A0A131YXS3 A0A131XE35 A0A1Q1M8K1 L7M1Z9 Q9W5W7 B4M9Y0
PDB
5X49
E-value=1.46888e-83,
Score=790
Ontologies
KEGG
GO
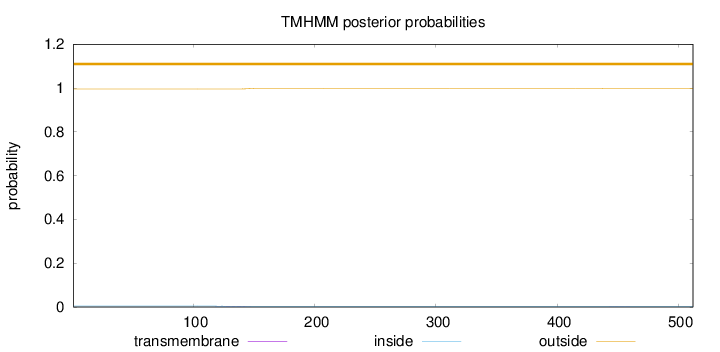
Topology
Length:
512
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0398700000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00421
outside
1 - 512
Population Genetic Test Statistics
Pi
274.323259
Theta
152.737948
Tajima's D
2.298599
CLR
0.233201
CSRT
0.92150392480376
Interpretation
Uncertain
