Gene
KWMTBOMO08672 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007935
Annotation
phosphoribosylaminoimidazole_carboxylase?_phosphoribosylaminoimidazole_succinocarboxamide_synthetase_[Bombyx_mori]
Full name
Multifunctional protein ADE2
Alternative Name
Protein adenosine-5
Location in the cell
Cytoplasmic Reliability : 2.894
Sequence
CDS
ATGTCTCATCCTAAACAAGTTGGACAATATAAATTGGGCAAACTTCTCATTGAGGGTAAGACCAAACAGGTATTCGATGTTCCGGACCAACCTGGGTACTGCTTGCTTCTTAACAAGGACAGAATCACCGCAGGCGATGGTGTCAAAGCCCACGATTTGGAGGGAAAGGCGGCTATTTCCAATCAGACAAATGCAAAAGTGTTCGAAATCTTAAAGTCTGCAGGAATCAAAACTGCCTTCGTTAAGATTGCCTCTGAAACTGCGTTCTTATCCAAAAAATGTGAGATGATCCCCATTGAATGGGTTACCCGCCGCCTTGCAACTGGCTCTTTCCTAAAAAGAAATCCTGGTGTACCAGAAGGCTTCAGGTTCACTCCTCCAAAACAAGAGACGTTCTTCAAGGATGACGCAAACCACGATCCCCAATGGTCAGAGGAGCAAATCATTTCAGCCAAATTCAATTATAACGGTCTTCTAATCGGTCGCGACGAGGTCGACTACATGAGAAAAGCTACGATTCTAATATTTGAAATCTTGGAGAAAGCTTGGGCTCTGCGTGACTGCGCACTTATTGACATGAAAATAGAATTCGGAGTCGATACAGAAGGTAGCATTGTATTGGCGGATGTCATCGATTCTGACTCTTGGAGATTGTGGCCTTCCGGTGATAAAAGATTAATGGTTGACAAACAGGTGTACCGGAACCTCACAACGGTGACTGCAGCTGATCTAGACACAGTCAAACGCAACTTTGCTTGGGTGAAAGACCAGCTTGATTTCTTGAAGCCGACGATCCACCACAAAGTTGTCGTCTTTATGGGTTCACCAGCCGACCAAGAACATTGTCAGAAAATAGCTAAAGCTGCCCGTGAACTTGGACTGGATGTCGACCTTCGCGTGACATCAGCCCACAAGGCCACTGAAGAAACCCTTCGCATTATGCAGCAATATGAAGATACGCATGGTGCTCTCGTGTTCATTGCTGTAGCAGGACGATCGAACGGACTCGGGCCCGTTCTTTCTGGGAACACTTCCTACCCTGTCATCAACTGCCCACCTCCTTCCGATAAACTTGTACAGGACATCTGGTCGTCCCTCTCCGTACCGTCTGGATTAGGATGCGCTACAGTAATTTATCCTGACAGTGCTGCACTAATGGCTGCTCAAATAATCGGCCTTCAGGACTATTTAGTGTGGGGACGTCTTAGATCCAAACAACTTGACATGGCACATTCACTAAGACAAGCGGATAAGAAGCTCAGGAACCAAACAGCTTAA
Protein
MSHPKQVGQYKLGKLLIEGKTKQVFDVPDQPGYCLLLNKDRITAGDGVKAHDLEGKAAISNQTNAKVFEILKSAGIKTAFVKIASETAFLSKKCEMIPIEWVTRRLATGSFLKRNPGVPEGFRFTPPKQETFFKDDANHDPQWSEEQIISAKFNYNGLLIGRDEVDYMRKATILIFEILEKAWALRDCALIDMKIEFGVDTEGSIVLADVIDSDSWRLWPSGDKRLMVDKQVYRNLTTVTAADLDTVKRNFAWVKDQLDFLKPTIHHKVVVFMGSPADQEHCQKIAKAARELGLDVDLRVTSAHKATEETLRIMQQYEDTHGALVFIAVAGRSNGLGPVLSGNTSYPVINCPPPSDKLVQDIWSSLSVPSGLGCATVIYPDSAALMAAQIIGLQDYLVWGRLRSKQLDMAHSLRQADKKLRNQTA
Summary
Catalytic Activity
5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate + ATP + L-aspartate = (2S)-2-[5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4-carboxamido]succinate + ADP + 2 H(+) + phosphate
5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate + H(+) = 5-amino-1-(5-phospho-beta-D-ribosyl)imidazole + CO2
5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate + H(+) = 5-amino-1-(5-phospho-beta-D-ribosyl)imidazole + CO2
Similarity
In the N-terminal section; belongs to the SAICAR synthetase family.
In the C-terminal section; belongs to the AIR carboxylase family. Class II subfamily.
In the C-terminal section; belongs to the AIR carboxylase family. Class II subfamily.
Keywords
ATP-binding
Complete proteome
Decarboxylase
Ligase
Lyase
Multifunctional enzyme
Nucleotide-binding
Purine biosynthesis
Reference proteome
Feature
chain Multifunctional protein ADE2
Uniprot
Q1HQ66
A0A437BXM1
A0A2H1WCU2
A0A2W1BMY5
A0A2A4IU77
S4PGW8
+ More
A0A212F8T0 A0A194PPN0 A1YM13 A0A1W4WUJ5 A0A1B0AP01 A0A1B0A4R8 A0A1Q3F2R0 D6X4R1 A0A1A9WEQ1 A0A034WLN4 Q17F22 A0A023ES15 A0A0A1WGL3 D3TM11 A0A1A9V290 A0A182GWZ0 A0A0C9RA29 T1E2N3 A0A182W508 B0WPD0 B3MS12 A0A0L0BSM2 A0A182R1A7 A0A1A9W5D7 W8BH16 A0A182SP00 A0A182LRQ2 A0A182P2A0 A0A0K8UGK4 A0A182YQ72 A0A336KFW7 A0A084VIX2 A0A182NAB1 A0A2M4A043 A0A2M4BP29 A0A182R7J1 A0A2M4BNQ5 T1DPD5 U5EW89 A0A2M3YXP0 B4L213 Q29GZ6 A0A2M3ZYZ5 A0A182FID9 A0A2M4BNI4 A0A0M4F9P8 A0A0A1WMV3 A0A2M3ZYG7 B4H0P3 A0A182JGL4 B4M6N9 W5JMN1 B4N1Z1 A0A1W4V7Q0 B4IGJ0 B4R3Z3 A0A182KCQ7 B3NXA0 A0A3B0K3H1 Q9I7S8 F0JAN1 A0A1J1HQD6 K7IQ46 U4UIX1 A0A1I8PRB4 T1P8U6 N6UJX4 A0A182X7Z1 A0A182UP35 A0A182KY39 A0A182UKZ6 A0A182HIC5 B4Q1Q3 A0A232FBA9 V5GWK5 A0A1I8NGM8 J3JUG4 Q7PRW1 A0A1B0DP59 E0VTD3 A0A067QNC4 A0A1Y1JZP7 B4JK15 A0A1B6EQT7 A0A0T6B715 A0A1L8DXY3 A0A1B0GK94 A0A1B6MDC7 A0A1B6C5I8 A0A161MQN1 A0A0A9WZ50 A0A2J7PC66 R4FNG8 A0A226DKZ0
A0A212F8T0 A0A194PPN0 A1YM13 A0A1W4WUJ5 A0A1B0AP01 A0A1B0A4R8 A0A1Q3F2R0 D6X4R1 A0A1A9WEQ1 A0A034WLN4 Q17F22 A0A023ES15 A0A0A1WGL3 D3TM11 A0A1A9V290 A0A182GWZ0 A0A0C9RA29 T1E2N3 A0A182W508 B0WPD0 B3MS12 A0A0L0BSM2 A0A182R1A7 A0A1A9W5D7 W8BH16 A0A182SP00 A0A182LRQ2 A0A182P2A0 A0A0K8UGK4 A0A182YQ72 A0A336KFW7 A0A084VIX2 A0A182NAB1 A0A2M4A043 A0A2M4BP29 A0A182R7J1 A0A2M4BNQ5 T1DPD5 U5EW89 A0A2M3YXP0 B4L213 Q29GZ6 A0A2M3ZYZ5 A0A182FID9 A0A2M4BNI4 A0A0M4F9P8 A0A0A1WMV3 A0A2M3ZYG7 B4H0P3 A0A182JGL4 B4M6N9 W5JMN1 B4N1Z1 A0A1W4V7Q0 B4IGJ0 B4R3Z3 A0A182KCQ7 B3NXA0 A0A3B0K3H1 Q9I7S8 F0JAN1 A0A1J1HQD6 K7IQ46 U4UIX1 A0A1I8PRB4 T1P8U6 N6UJX4 A0A182X7Z1 A0A182UP35 A0A182KY39 A0A182UKZ6 A0A182HIC5 B4Q1Q3 A0A232FBA9 V5GWK5 A0A1I8NGM8 J3JUG4 Q7PRW1 A0A1B0DP59 E0VTD3 A0A067QNC4 A0A1Y1JZP7 B4JK15 A0A1B6EQT7 A0A0T6B715 A0A1L8DXY3 A0A1B0GK94 A0A1B6MDC7 A0A1B6C5I8 A0A161MQN1 A0A0A9WZ50 A0A2J7PC66 R4FNG8 A0A226DKZ0
Pubmed
19121390
23553965
28756777
23622113
22118469
26354079
+ More
18362917 19820115 25348373 17510324 24945155 25830018 20353571 26483478 24330624 17994087 26108605 24495485 25244985 24438588 15632085 20920257 23761445 10757766 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 23537049 20966253 17550304 28648823 25315136 22516182 12364791 14747013 17210077 20566863 24845553 28004739 25401762
18362917 19820115 25348373 17510324 24945155 25830018 20353571 26483478 24330624 17994087 26108605 24495485 25244985 24438588 15632085 20920257 23761445 10757766 10731132 12537572 12537569 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 23537049 20966253 17550304 28648823 25315136 22516182 12364791 14747013 17210077 20566863 24845553 28004739 25401762
EMBL
BABH01003213
DQ443186
ABF51275.1
RSAL01000001
RVE55188.1
ODYU01007812
+ More
SOQ50895.1 KZ150060 PZC74236.1 NWSH01006302 PCG63527.1 GAIX01003472 JAA89088.1 AGBW02009681 OWR50123.1 KQ459596 KPI95401.1 EF105371 ABM05496.1 JXJN01001088 JXJN01001089 JXJN01001090 GFDL01013226 JAV21819.1 KQ971380 EEZ97680.1 GAKP01003383 JAC55569.1 CH477277 EAT45109.1 GAPW01001580 JAC12018.1 GBXI01016652 GBXI01011738 JAC97639.1 JAD02554.1 CCAG010023143 EZ422463 ADD18739.1 JXUM01094256 KQ564116 KXJ72751.1 GBYB01005095 JAG74862.1 GALA01001125 JAA93727.1 DS232022 EDS32305.1 CH902622 EDV34567.1 JRES01001419 KNC23080.1 AXCN02001415 GAMC01005990 JAC00566.1 AXCM01007375 GDHF01026517 GDHF01018197 GDHF01001433 JAI25797.1 JAI34117.1 JAI50881.1 UFQS01000116 UFQT01000116 SSW99960.1 SSX20340.1 ATLV01013412 KE524855 KFB37916.1 GGFK01000800 MBW34121.1 GGFJ01005560 MBW54701.1 GGFJ01005501 MBW54642.1 GAMD01003434 JAA98156.1 GANO01001535 JAB58336.1 GGFM01000257 MBW21008.1 CH933810 EDW07734.1 CH379064 EAL31963.1 GGFK01000371 MBW33692.1 GGFJ01005505 MBW54646.1 CP012528 ALC48995.1 GBXI01014492 JAC99799.1 GGFK01000282 MBW33603.1 CH479200 EDW29838.1 CH940653 EDW62456.1 ADMH02000959 ETN64568.1 CH963925 EDW78380.1 CH480836 EDW48940.1 CM000366 EDX17760.1 CH954180 EDV47272.1 OUUW01000016 SPP88775.1 AF102579 AF102580 AE014298 AY095520 AAF06356.1 AAG22346.2 AAM12252.1 BT126073 ADY17772.1 AGB95335.1 CVRI01000013 CRK89596.1 KB632192 ERL89880.1 KA645107 AFP59736.1 APGK01017070 APGK01017071 KB739995 ENN82030.1 APCN01003927 CM000162 EDX01494.1 NNAY01000571 OXU27617.1 GALX01002379 JAB66087.1 BT126877 AEE61839.1 AAAB01008846 EAA06482.5 AJVK01007915 DS235762 EEB16639.1 KK853205 KDR09851.1 GEZM01099550 JAV53280.1 CH916370 EDV99917.1 GECZ01029501 JAS40268.1 LJIG01009441 KRT83073.1 GFDF01002810 JAV11274.1 AJWK01028576 GEBQ01006088 JAT33889.1 GEDC01028551 JAS08747.1 GEMB01002231 JAS00949.1 GBHO01030913 GBRD01001436 JAG12691.1 JAG64385.1 NEVH01027067 PNF13929.1 ACPB03003540 GAHY01001299 JAA76211.1 LNIX01000017 OXA45658.1
SOQ50895.1 KZ150060 PZC74236.1 NWSH01006302 PCG63527.1 GAIX01003472 JAA89088.1 AGBW02009681 OWR50123.1 KQ459596 KPI95401.1 EF105371 ABM05496.1 JXJN01001088 JXJN01001089 JXJN01001090 GFDL01013226 JAV21819.1 KQ971380 EEZ97680.1 GAKP01003383 JAC55569.1 CH477277 EAT45109.1 GAPW01001580 JAC12018.1 GBXI01016652 GBXI01011738 JAC97639.1 JAD02554.1 CCAG010023143 EZ422463 ADD18739.1 JXUM01094256 KQ564116 KXJ72751.1 GBYB01005095 JAG74862.1 GALA01001125 JAA93727.1 DS232022 EDS32305.1 CH902622 EDV34567.1 JRES01001419 KNC23080.1 AXCN02001415 GAMC01005990 JAC00566.1 AXCM01007375 GDHF01026517 GDHF01018197 GDHF01001433 JAI25797.1 JAI34117.1 JAI50881.1 UFQS01000116 UFQT01000116 SSW99960.1 SSX20340.1 ATLV01013412 KE524855 KFB37916.1 GGFK01000800 MBW34121.1 GGFJ01005560 MBW54701.1 GGFJ01005501 MBW54642.1 GAMD01003434 JAA98156.1 GANO01001535 JAB58336.1 GGFM01000257 MBW21008.1 CH933810 EDW07734.1 CH379064 EAL31963.1 GGFK01000371 MBW33692.1 GGFJ01005505 MBW54646.1 CP012528 ALC48995.1 GBXI01014492 JAC99799.1 GGFK01000282 MBW33603.1 CH479200 EDW29838.1 CH940653 EDW62456.1 ADMH02000959 ETN64568.1 CH963925 EDW78380.1 CH480836 EDW48940.1 CM000366 EDX17760.1 CH954180 EDV47272.1 OUUW01000016 SPP88775.1 AF102579 AF102580 AE014298 AY095520 AAF06356.1 AAG22346.2 AAM12252.1 BT126073 ADY17772.1 AGB95335.1 CVRI01000013 CRK89596.1 KB632192 ERL89880.1 KA645107 AFP59736.1 APGK01017070 APGK01017071 KB739995 ENN82030.1 APCN01003927 CM000162 EDX01494.1 NNAY01000571 OXU27617.1 GALX01002379 JAB66087.1 BT126877 AEE61839.1 AAAB01008846 EAA06482.5 AJVK01007915 DS235762 EEB16639.1 KK853205 KDR09851.1 GEZM01099550 JAV53280.1 CH916370 EDV99917.1 GECZ01029501 JAS40268.1 LJIG01009441 KRT83073.1 GFDF01002810 JAV11274.1 AJWK01028576 GEBQ01006088 JAT33889.1 GEDC01028551 JAS08747.1 GEMB01002231 JAS00949.1 GBHO01030913 GBRD01001436 JAG12691.1 JAG64385.1 NEVH01027067 PNF13929.1 ACPB03003540 GAHY01001299 JAA76211.1 LNIX01000017 OXA45658.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000192223
+ More
UP000092460 UP000092445 UP000007266 UP000091820 UP000008820 UP000092444 UP000078200 UP000069940 UP000249989 UP000075920 UP000002320 UP000007801 UP000037069 UP000075886 UP000075901 UP000075883 UP000075885 UP000076408 UP000030765 UP000075884 UP000075900 UP000009192 UP000001819 UP000069272 UP000092553 UP000008744 UP000075880 UP000008792 UP000000673 UP000007798 UP000192221 UP000001292 UP000000304 UP000075881 UP000008711 UP000268350 UP000000803 UP000183832 UP000002358 UP000030742 UP000095300 UP000019118 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000002282 UP000215335 UP000095301 UP000007062 UP000092462 UP000009046 UP000027135 UP000001070 UP000092461 UP000235965 UP000015103 UP000198287
UP000092460 UP000092445 UP000007266 UP000091820 UP000008820 UP000092444 UP000078200 UP000069940 UP000249989 UP000075920 UP000002320 UP000007801 UP000037069 UP000075886 UP000075901 UP000075883 UP000075885 UP000076408 UP000030765 UP000075884 UP000075900 UP000009192 UP000001819 UP000069272 UP000092553 UP000008744 UP000075880 UP000008792 UP000000673 UP000007798 UP000192221 UP000001292 UP000000304 UP000075881 UP000008711 UP000268350 UP000000803 UP000183832 UP000002358 UP000030742 UP000095300 UP000019118 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000002282 UP000215335 UP000095301 UP000007062 UP000092462 UP000009046 UP000027135 UP000001070 UP000092461 UP000235965 UP000015103 UP000198287
Interpro
Gene 3D
CDD
ProteinModelPortal
Q1HQ66
A0A437BXM1
A0A2H1WCU2
A0A2W1BMY5
A0A2A4IU77
S4PGW8
+ More
A0A212F8T0 A0A194PPN0 A1YM13 A0A1W4WUJ5 A0A1B0AP01 A0A1B0A4R8 A0A1Q3F2R0 D6X4R1 A0A1A9WEQ1 A0A034WLN4 Q17F22 A0A023ES15 A0A0A1WGL3 D3TM11 A0A1A9V290 A0A182GWZ0 A0A0C9RA29 T1E2N3 A0A182W508 B0WPD0 B3MS12 A0A0L0BSM2 A0A182R1A7 A0A1A9W5D7 W8BH16 A0A182SP00 A0A182LRQ2 A0A182P2A0 A0A0K8UGK4 A0A182YQ72 A0A336KFW7 A0A084VIX2 A0A182NAB1 A0A2M4A043 A0A2M4BP29 A0A182R7J1 A0A2M4BNQ5 T1DPD5 U5EW89 A0A2M3YXP0 B4L213 Q29GZ6 A0A2M3ZYZ5 A0A182FID9 A0A2M4BNI4 A0A0M4F9P8 A0A0A1WMV3 A0A2M3ZYG7 B4H0P3 A0A182JGL4 B4M6N9 W5JMN1 B4N1Z1 A0A1W4V7Q0 B4IGJ0 B4R3Z3 A0A182KCQ7 B3NXA0 A0A3B0K3H1 Q9I7S8 F0JAN1 A0A1J1HQD6 K7IQ46 U4UIX1 A0A1I8PRB4 T1P8U6 N6UJX4 A0A182X7Z1 A0A182UP35 A0A182KY39 A0A182UKZ6 A0A182HIC5 B4Q1Q3 A0A232FBA9 V5GWK5 A0A1I8NGM8 J3JUG4 Q7PRW1 A0A1B0DP59 E0VTD3 A0A067QNC4 A0A1Y1JZP7 B4JK15 A0A1B6EQT7 A0A0T6B715 A0A1L8DXY3 A0A1B0GK94 A0A1B6MDC7 A0A1B6C5I8 A0A161MQN1 A0A0A9WZ50 A0A2J7PC66 R4FNG8 A0A226DKZ0
A0A212F8T0 A0A194PPN0 A1YM13 A0A1W4WUJ5 A0A1B0AP01 A0A1B0A4R8 A0A1Q3F2R0 D6X4R1 A0A1A9WEQ1 A0A034WLN4 Q17F22 A0A023ES15 A0A0A1WGL3 D3TM11 A0A1A9V290 A0A182GWZ0 A0A0C9RA29 T1E2N3 A0A182W508 B0WPD0 B3MS12 A0A0L0BSM2 A0A182R1A7 A0A1A9W5D7 W8BH16 A0A182SP00 A0A182LRQ2 A0A182P2A0 A0A0K8UGK4 A0A182YQ72 A0A336KFW7 A0A084VIX2 A0A182NAB1 A0A2M4A043 A0A2M4BP29 A0A182R7J1 A0A2M4BNQ5 T1DPD5 U5EW89 A0A2M3YXP0 B4L213 Q29GZ6 A0A2M3ZYZ5 A0A182FID9 A0A2M4BNI4 A0A0M4F9P8 A0A0A1WMV3 A0A2M3ZYG7 B4H0P3 A0A182JGL4 B4M6N9 W5JMN1 B4N1Z1 A0A1W4V7Q0 B4IGJ0 B4R3Z3 A0A182KCQ7 B3NXA0 A0A3B0K3H1 Q9I7S8 F0JAN1 A0A1J1HQD6 K7IQ46 U4UIX1 A0A1I8PRB4 T1P8U6 N6UJX4 A0A182X7Z1 A0A182UP35 A0A182KY39 A0A182UKZ6 A0A182HIC5 B4Q1Q3 A0A232FBA9 V5GWK5 A0A1I8NGM8 J3JUG4 Q7PRW1 A0A1B0DP59 E0VTD3 A0A067QNC4 A0A1Y1JZP7 B4JK15 A0A1B6EQT7 A0A0T6B715 A0A1L8DXY3 A0A1B0GK94 A0A1B6MDC7 A0A1B6C5I8 A0A161MQN1 A0A0A9WZ50 A0A2J7PC66 R4FNG8 A0A226DKZ0
PDB
4JA0
E-value=0,
Score=2269
Ontologies
PATHWAY
GO
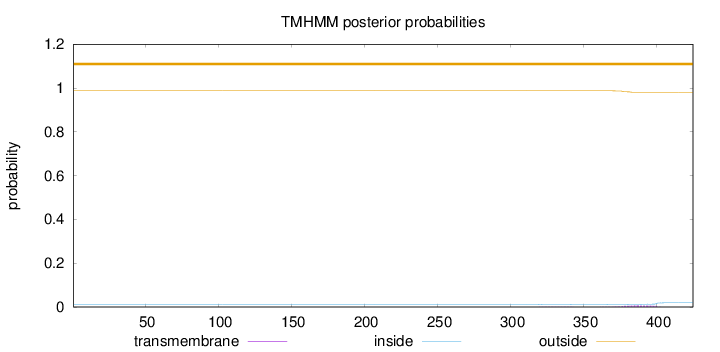
Topology
Length:
425
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1767
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01235
outside
1 - 425
Population Genetic Test Statistics
Pi
255.863641
Theta
185.958823
Tajima's D
0.959667
CLR
12.090716
CSRT
0.645767711614419
Interpretation
Uncertain
