Gene
KWMTBOMO08656
Pre Gene Modal
BGIBMGA007944
Annotation
PREDICTED:_iron-sulfur_protein_NUBPL_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.454
Sequence
CDS
ATGACCCTACAGCGTAGGTTTCTCCGCTTCTCTAGTAGTTATGTGTTCAGTACCAGGATGAACACGATAGCACCATGCCTTCAAACAGTAAGATTTAACCACAGTAAAAGTGACATTATGGATCATAGAGCCAAAGTTATGAGCAAAGGGTTACCAGAGAAAAAGCCCCTACCTGGTGTAAAGAGTATCATACTTGTAGCTTCAGGGAAAGGAGGTGTTGGAAAAACTACCACAGCTGTAAACCTAGCATGTGCTATGAAAGTAATAGAACCGGACAAGGAAATAGGGCTCCTAGATGCTGATGTGTTTGGTCCTTCAGTACCATTGATGATGAATATCAGTGGAGAACCAATGCTTAATGACGATCATCTCATTGAACCTTTACTTAATTATGGTGTTAAATGTATGTCAATGGGTCTATTAGTGTCTGGCGAGAATGCTGTAGTGTGGCGCGGCTTAATGGTGATGCAAGCACTGGAACGTCTGACGCGACACGTGGCGTGGGGTCCACTTGACTGTCTCGTTGTAGATACGCCGCCTGGTACAGGCGATACACATTTATCACTTGCGCAAAACTTGCCTTTAGATGGAGCAATAGTAGTAACAACACCCCAATCAGCCGCATTGCAAGTAACTAAAAGAGGCATAAATATGTTCGAGAAATTAAAGGTTCCTATTATAGATATGGGTCTTGAGATTATTGAAAGTTTTGAAGTAGATCAAAATATGTCAGATTGTATAAACAGCGGGAAGCCAGCGATTTACGCCCTCCCAGACAGCATACACGCAGAGAAATACAGACACCTTGCTAATAAAGTTTTTAAATATATAGATGATAAGGAAAACCACAGAGCAAAAGCCAAAGAACAATGA
Protein
MTLQRRFLRFSSSYVFSTRMNTIAPCLQTVRFNHSKSDIMDHRAKVMSKGLPEKKPLPGVKSIILVASGKGGVGKTTTAVNLACAMKVIEPDKEIGLLDADVFGPSVPLMMNISGEPMLNDDHLIEPLLNYGVKCMSMGLLVSGENAVVWRGLMVMQALERLTRHVAWGPLDCLVVDTPPGTGDTHLSLAQNLPLDGAIVVTTPQSAALQVTKRGINMFEKLKVPIIDMGLEIIESFEVDQNMSDCINSGKPAIYALPDSIHAEKYRHLANKVFKYIDDKENHRAKAKEQ
Summary
Uniprot
H9JEJ7
A0A2H1VA84
A0A2A4K6W2
A0A3S2MAK0
S4NXE5
A0A194PPZ5
+ More
A0A212F8S8 A0A0L7R500 A0A0M8ZQP7 A0A2A3E2C7 A0A310SND5 A0A1B6GHX5 A0A2J7RQ48 A0A1B6KHB6 J3JXD2 A0A088AL15 D6WCQ3 A0A2J7RQ36 A0A195E731 A0A154P7D6 A0A026VSI1 A0A151XBA4 E2AQH6 F4WTP4 A0A195F219 A0A151I196 A0A158NG45 A0A2A3E1L3 E2BIV9 A0A195CI33 U4ULW9 A0A1B6KMF2 A0A1W4WTN0 A0A1W4WHG9 A0A067RDM6 K7IQ51 A0A2H8TEZ5 A0A2P8XXX2 T1I708 N6U464 A0A1B6HZW7 A0A1I8MEY5 E0VGD8 L7M289 A0A0P4VLX9 A0A1J1HWA8 A0A1B6LPP1 A0A226ENJ8 N6U6X3 A0A1I8Q873 F2UF70 A0A034WH45 M4A4U7 G3P9Q3 A0A0L0C3S2 A0A034WD03 A0A0K8R6B0 A0A0A9YEV5 A0A131Y475 A0A1B6JYR6 C4WWF6 A0A3B3BFU2 A0A182QI80 A0A0K8UD85 W8BNR0 A0A336LFZ4 A0A232FAV9 A0A3P9N0W1 B4MU69 H2S6F4 A0A336MZL2 T1IY64 A0A0V0GBX3 A0A3Q2EER8 A0A147AJD2 A0A131Z0Y0 B4P6C0 A0A1S3IPK2 V3ZPY6 A0A3B3Z124 A0A3P9KPC9 W5MTG8 A0A0R1DQH6 A0A3B3UYV6 A0A3Q1G2K9 A0A3B4EDV8 A0A146XHX1 A0A3Q3IVN6 A0A087Y763 H2MUZ8 A0A3P9HI14 A0A2R7VZY5 Q28I04 Q16JY4 B3NKN6 A0A3Q3L7A0 H0X515
A0A212F8S8 A0A0L7R500 A0A0M8ZQP7 A0A2A3E2C7 A0A310SND5 A0A1B6GHX5 A0A2J7RQ48 A0A1B6KHB6 J3JXD2 A0A088AL15 D6WCQ3 A0A2J7RQ36 A0A195E731 A0A154P7D6 A0A026VSI1 A0A151XBA4 E2AQH6 F4WTP4 A0A195F219 A0A151I196 A0A158NG45 A0A2A3E1L3 E2BIV9 A0A195CI33 U4ULW9 A0A1B6KMF2 A0A1W4WTN0 A0A1W4WHG9 A0A067RDM6 K7IQ51 A0A2H8TEZ5 A0A2P8XXX2 T1I708 N6U464 A0A1B6HZW7 A0A1I8MEY5 E0VGD8 L7M289 A0A0P4VLX9 A0A1J1HWA8 A0A1B6LPP1 A0A226ENJ8 N6U6X3 A0A1I8Q873 F2UF70 A0A034WH45 M4A4U7 G3P9Q3 A0A0L0C3S2 A0A034WD03 A0A0K8R6B0 A0A0A9YEV5 A0A131Y475 A0A1B6JYR6 C4WWF6 A0A3B3BFU2 A0A182QI80 A0A0K8UD85 W8BNR0 A0A336LFZ4 A0A232FAV9 A0A3P9N0W1 B4MU69 H2S6F4 A0A336MZL2 T1IY64 A0A0V0GBX3 A0A3Q2EER8 A0A147AJD2 A0A131Z0Y0 B4P6C0 A0A1S3IPK2 V3ZPY6 A0A3B3Z124 A0A3P9KPC9 W5MTG8 A0A0R1DQH6 A0A3B3UYV6 A0A3Q1G2K9 A0A3B4EDV8 A0A146XHX1 A0A3Q3IVN6 A0A087Y763 H2MUZ8 A0A3P9HI14 A0A2R7VZY5 Q28I04 Q16JY4 B3NKN6 A0A3Q3L7A0 H0X515
Pubmed
19121390
23622113
26354079
22118469
22516182
18362917
+ More
19820115 24508170 30249741 20798317 21719571 21347285 23537049 24845553 20075255 29403074 25315136 20566863 25576852 27129103 25348373 23542700 26108605 25401762 26823975 29451363 24495485 28648823 17994087 21551351 26830274 17550304 23254933 17554307 17510324
19820115 24508170 30249741 20798317 21719571 21347285 23537049 24845553 20075255 29403074 25315136 20566863 25576852 27129103 25348373 23542700 26108605 25401762 26823975 29451363 24495485 28648823 17994087 21551351 26830274 17550304 23254933 17554307 17510324
EMBL
BABH01003231
ODYU01001486
SOQ37748.1
NWSH01000087
PCG79756.1
RSAL01000001
+ More
RVE55202.1 GAIX01009069 JAA83491.1 KQ459596 KPI95387.1 AGBW02009681 OWR50137.1 KQ414654 KOC65923.1 KQ435989 KOX67683.1 KZ288445 PBC25634.1 KQ761783 OAD57028.1 GECZ01007758 JAS62011.1 NEVH01001348 PNF42954.1 GEBQ01029119 JAT10858.1 BT127901 AEE62863.1 KQ971317 EEZ98808.2 PNF42955.1 KQ979568 KYN21005.1 KQ434831 KZC07839.1 KK108395 QOIP01000006 EZA46718.1 RLU21159.1 KQ982335 KYQ57661.1 GL441764 EFN64324.1 GL888341 EGI62465.1 KQ981864 KYN34139.1 KQ976580 KYM79958.1 ADTU01014962 PBC25633.1 GL448535 EFN84391.1 KQ977721 KYN00368.1 KB632379 ERL94117.1 GEBQ01027603 JAT12374.1 KK852703 KDR18089.1 GFXV01000840 MBW12645.1 PYGN01001182 PSN36836.1 ACPB03007038 APGK01040365 KB740979 ENN76395.1 GECU01027490 JAS80216.1 DS235138 EEB12444.1 GACK01007052 JAA57982.1 GDKW01003146 JAI53449.1 CVRI01000020 CRK90798.1 GEBQ01014305 JAT25672.1 LNIX01000003 OXA58788.1 ENN76396.1 GL832971 EGD75270.1 GAKP01005492 JAC53460.1 JRES01000940 KNC26988.1 GAKP01005491 JAC53461.1 GADI01007217 JAA66591.1 GBHO01013991 GBHO01013990 GBRD01006039 GDHC01010206 JAG29613.1 JAG29614.1 JAG59782.1 JAQ08423.1 GEFM01002488 JAP73308.1 GECU01003653 JAT04054.1 ABLF02029830 AK342039 BAH72226.1 AXCN02000732 GDHF01027786 JAI24528.1 GAMC01011514 JAB95041.1 UFQS01003316 UFQT01003316 SSX15485.1 SSX34851.1 NNAY01000571 OXU27620.1 CH963852 EDW75658.1 UFQT01003132 SSX34619.1 JH431671 GECL01000783 JAP05341.1 GCES01008091 JAR78232.1 GEDV01003468 JAP85089.1 CM000158 EDW89877.1 KB203470 ESO84565.1 AHAT01003856 KRJ98765.1 GCES01044605 JAR41718.1 AYCK01021768 AYCK01021769 KK854205 PTY13057.1 CR760654 CAJ81382.1 CH477986 EAT34599.1 CH954179 EDV54340.1 AAQR03086288 AAQR03086289 AAQR03086290 AAQR03086291 AAQR03086292 AAQR03086293 AAQR03086294 AAQR03086295 AAQR03086296 AAQR03086297
RVE55202.1 GAIX01009069 JAA83491.1 KQ459596 KPI95387.1 AGBW02009681 OWR50137.1 KQ414654 KOC65923.1 KQ435989 KOX67683.1 KZ288445 PBC25634.1 KQ761783 OAD57028.1 GECZ01007758 JAS62011.1 NEVH01001348 PNF42954.1 GEBQ01029119 JAT10858.1 BT127901 AEE62863.1 KQ971317 EEZ98808.2 PNF42955.1 KQ979568 KYN21005.1 KQ434831 KZC07839.1 KK108395 QOIP01000006 EZA46718.1 RLU21159.1 KQ982335 KYQ57661.1 GL441764 EFN64324.1 GL888341 EGI62465.1 KQ981864 KYN34139.1 KQ976580 KYM79958.1 ADTU01014962 PBC25633.1 GL448535 EFN84391.1 KQ977721 KYN00368.1 KB632379 ERL94117.1 GEBQ01027603 JAT12374.1 KK852703 KDR18089.1 GFXV01000840 MBW12645.1 PYGN01001182 PSN36836.1 ACPB03007038 APGK01040365 KB740979 ENN76395.1 GECU01027490 JAS80216.1 DS235138 EEB12444.1 GACK01007052 JAA57982.1 GDKW01003146 JAI53449.1 CVRI01000020 CRK90798.1 GEBQ01014305 JAT25672.1 LNIX01000003 OXA58788.1 ENN76396.1 GL832971 EGD75270.1 GAKP01005492 JAC53460.1 JRES01000940 KNC26988.1 GAKP01005491 JAC53461.1 GADI01007217 JAA66591.1 GBHO01013991 GBHO01013990 GBRD01006039 GDHC01010206 JAG29613.1 JAG29614.1 JAG59782.1 JAQ08423.1 GEFM01002488 JAP73308.1 GECU01003653 JAT04054.1 ABLF02029830 AK342039 BAH72226.1 AXCN02000732 GDHF01027786 JAI24528.1 GAMC01011514 JAB95041.1 UFQS01003316 UFQT01003316 SSX15485.1 SSX34851.1 NNAY01000571 OXU27620.1 CH963852 EDW75658.1 UFQT01003132 SSX34619.1 JH431671 GECL01000783 JAP05341.1 GCES01008091 JAR78232.1 GEDV01003468 JAP85089.1 CM000158 EDW89877.1 KB203470 ESO84565.1 AHAT01003856 KRJ98765.1 GCES01044605 JAR41718.1 AYCK01021768 AYCK01021769 KK854205 PTY13057.1 CR760654 CAJ81382.1 CH477986 EAT34599.1 CH954179 EDV54340.1 AAQR03086288 AAQR03086289 AAQR03086290 AAQR03086291 AAQR03086292 AAQR03086293 AAQR03086294 AAQR03086295 AAQR03086296 AAQR03086297
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000053825
+ More
UP000053105 UP000242457 UP000235965 UP000005203 UP000007266 UP000078492 UP000076502 UP000053097 UP000279307 UP000075809 UP000000311 UP000007755 UP000078541 UP000078540 UP000005205 UP000008237 UP000078542 UP000030742 UP000192223 UP000027135 UP000002358 UP000245037 UP000015103 UP000019118 UP000095301 UP000009046 UP000183832 UP000198287 UP000095300 UP000007799 UP000002852 UP000007635 UP000037069 UP000007819 UP000261560 UP000075886 UP000215335 UP000242638 UP000007798 UP000005226 UP000265020 UP000002282 UP000085678 UP000030746 UP000261480 UP000265180 UP000018468 UP000261500 UP000257200 UP000261440 UP000261600 UP000028760 UP000001038 UP000265200 UP000008820 UP000008711 UP000261660 UP000005225
UP000053105 UP000242457 UP000235965 UP000005203 UP000007266 UP000078492 UP000076502 UP000053097 UP000279307 UP000075809 UP000000311 UP000007755 UP000078541 UP000078540 UP000005205 UP000008237 UP000078542 UP000030742 UP000192223 UP000027135 UP000002358 UP000245037 UP000015103 UP000019118 UP000095301 UP000009046 UP000183832 UP000198287 UP000095300 UP000007799 UP000002852 UP000007635 UP000037069 UP000007819 UP000261560 UP000075886 UP000215335 UP000242638 UP000007798 UP000005226 UP000265020 UP000002282 UP000085678 UP000030746 UP000261480 UP000265180 UP000018468 UP000261500 UP000257200 UP000261440 UP000261600 UP000028760 UP000001038 UP000265200 UP000008820 UP000008711 UP000261660 UP000005225
PRIDE
Interpro
CDD
ProteinModelPortal
H9JEJ7
A0A2H1VA84
A0A2A4K6W2
A0A3S2MAK0
S4NXE5
A0A194PPZ5
+ More
A0A212F8S8 A0A0L7R500 A0A0M8ZQP7 A0A2A3E2C7 A0A310SND5 A0A1B6GHX5 A0A2J7RQ48 A0A1B6KHB6 J3JXD2 A0A088AL15 D6WCQ3 A0A2J7RQ36 A0A195E731 A0A154P7D6 A0A026VSI1 A0A151XBA4 E2AQH6 F4WTP4 A0A195F219 A0A151I196 A0A158NG45 A0A2A3E1L3 E2BIV9 A0A195CI33 U4ULW9 A0A1B6KMF2 A0A1W4WTN0 A0A1W4WHG9 A0A067RDM6 K7IQ51 A0A2H8TEZ5 A0A2P8XXX2 T1I708 N6U464 A0A1B6HZW7 A0A1I8MEY5 E0VGD8 L7M289 A0A0P4VLX9 A0A1J1HWA8 A0A1B6LPP1 A0A226ENJ8 N6U6X3 A0A1I8Q873 F2UF70 A0A034WH45 M4A4U7 G3P9Q3 A0A0L0C3S2 A0A034WD03 A0A0K8R6B0 A0A0A9YEV5 A0A131Y475 A0A1B6JYR6 C4WWF6 A0A3B3BFU2 A0A182QI80 A0A0K8UD85 W8BNR0 A0A336LFZ4 A0A232FAV9 A0A3P9N0W1 B4MU69 H2S6F4 A0A336MZL2 T1IY64 A0A0V0GBX3 A0A3Q2EER8 A0A147AJD2 A0A131Z0Y0 B4P6C0 A0A1S3IPK2 V3ZPY6 A0A3B3Z124 A0A3P9KPC9 W5MTG8 A0A0R1DQH6 A0A3B3UYV6 A0A3Q1G2K9 A0A3B4EDV8 A0A146XHX1 A0A3Q3IVN6 A0A087Y763 H2MUZ8 A0A3P9HI14 A0A2R7VZY5 Q28I04 Q16JY4 B3NKN6 A0A3Q3L7A0 H0X515
A0A212F8S8 A0A0L7R500 A0A0M8ZQP7 A0A2A3E2C7 A0A310SND5 A0A1B6GHX5 A0A2J7RQ48 A0A1B6KHB6 J3JXD2 A0A088AL15 D6WCQ3 A0A2J7RQ36 A0A195E731 A0A154P7D6 A0A026VSI1 A0A151XBA4 E2AQH6 F4WTP4 A0A195F219 A0A151I196 A0A158NG45 A0A2A3E1L3 E2BIV9 A0A195CI33 U4ULW9 A0A1B6KMF2 A0A1W4WTN0 A0A1W4WHG9 A0A067RDM6 K7IQ51 A0A2H8TEZ5 A0A2P8XXX2 T1I708 N6U464 A0A1B6HZW7 A0A1I8MEY5 E0VGD8 L7M289 A0A0P4VLX9 A0A1J1HWA8 A0A1B6LPP1 A0A226ENJ8 N6U6X3 A0A1I8Q873 F2UF70 A0A034WH45 M4A4U7 G3P9Q3 A0A0L0C3S2 A0A034WD03 A0A0K8R6B0 A0A0A9YEV5 A0A131Y475 A0A1B6JYR6 C4WWF6 A0A3B3BFU2 A0A182QI80 A0A0K8UD85 W8BNR0 A0A336LFZ4 A0A232FAV9 A0A3P9N0W1 B4MU69 H2S6F4 A0A336MZL2 T1IY64 A0A0V0GBX3 A0A3Q2EER8 A0A147AJD2 A0A131Z0Y0 B4P6C0 A0A1S3IPK2 V3ZPY6 A0A3B3Z124 A0A3P9KPC9 W5MTG8 A0A0R1DQH6 A0A3B3UYV6 A0A3Q1G2K9 A0A3B4EDV8 A0A146XHX1 A0A3Q3IVN6 A0A087Y763 H2MUZ8 A0A3P9HI14 A0A2R7VZY5 Q28I04 Q16JY4 B3NKN6 A0A3Q3L7A0 H0X515
PDB
2PH1
E-value=1.73213e-23,
Score=269
Ontologies
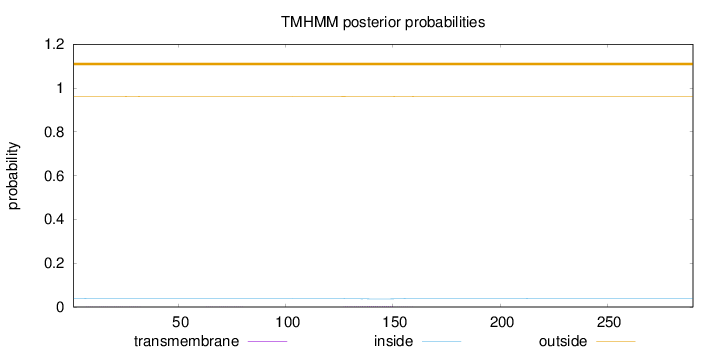
Topology
Length:
290
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0812299999999999
Exp number, first 60 AAs:
0.01074
Total prob of N-in:
0.03931
outside
1 - 290
Population Genetic Test Statistics
Pi
162.51009
Theta
14.095161
Tajima's D
0.558869
CLR
1.309975
CSRT
0.543422828858557
Interpretation
Uncertain
