Gene
KWMTBOMO08653
Pre Gene Modal
BGIBMGA007945
Annotation
hypothetical_protein_KGM_17711_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 1.962
Sequence
CDS
ATGATGACTGAAATCGATGAGTTCAATCAATACCAGAAGTCTTCTAAAACTGGAAAACAGCATAATGTTTTACCGATATGGGGTAACGAACAGACCATGAACTTGAATCCACTAATATTAGCCAATATTCAAGGCTCTAGCTACTTCAAAGTGCACCTGTTCAAGTTGAAGACCTACCATGAGGTGGTAGACGAAATATATTATCAGGTGAAACACCTTGAGCCGTGGGAGCGTGGTAGCCGCAAGACATCAGGCCAGACCGGCATGTGTGGCGGCGTACGAGGTGTGGGTGCTGGTGGAATTGTGTCCACTGCTTTCTGTCTCCTCTACAAGCTGTACACTCTCCGGTTGACACGAAAACAAGTTAACGGGTTGTTGCAACACACTGACTCTCCATATATCAGAGCACTGGGCTTCATGTACATTCGATATACACAACCACCAGCTGATCTATTTGACTGGTATGTGGATTACTTTGAAGATGAAGAAGAGGTTGACCCCCGTGCTGGAGGTGGTGCATCCACAACTATTGGAGCACTAGTACGACAAATGCTTGTAAAACTTGACTGGTTTAGCACGTTGTTTCCAAGGATACCTGTTCCCATCCAGAAACAGATTGAACAGAAATTAGCAGAGCACAATCGCAGTATTGCACCAACAAAGCCGGCCCCAGCGGCAAGGGCCTCCTCCTACAACCCAGGACGGATGGAATCTGATCGCCGCGACCACGACGACCGCGACCGCAGGGACTACGACGACCGCGACCGCAGGGACTACGATGACCGCGATCGCAGGGACTACGCCGATAGGAATGGAGGTGACAGGTAG
Protein
MMTEIDEFNQYQKSSKTGKQHNVLPIWGNEQTMNLNPLILANIQGSSYFKVHLFKLKTYHEVVDEIYYQVKHLEPWERGSRKTSGQTGMCGGVRGVGAGGIVSTAFCLLYKLYTLRLTRKQVNGLLQHTDSPYIRALGFMYIRYTQPPADLFDWYVDYFEDEEEVDPRAGGGASTTIGALVRQMLVKLDWFSTLFPRIPVPIQKQIEQKLAEHNRSIAPTKPAPAARASSYNPGRMESDRRDHDDRDRRDYDDRDRRDYDDRDRRDYADRNGGDR
Summary
Uniprot
H9JEJ8
A0A3S2LJF9
A0A212F8U1
A0A2A4K7U1
A0A2H1VAC8
A0A194PRK7
+ More
A0A0N1IAW4 S4PMT8 A0A0K8VK64 A0A034WTL0 A0A0A1XNR8 W8C0X7 A0A0L0BU41 A0A0K8TRH0 B0WBK2 A0A1A9WZE4 A0A1A9XBP0 A0A1B0ATG2 A0A1I8N7Q3 A0A1I8QBR4 A0A182Q8Z3 A0A182NVL4 A0A182SYW4 A0A084W1A0 A0A182YI84 A0A2M4CW95 A0A182IXY3 A0A1A9ZEP2 A0A1A9VCQ1 A0A2M4CW98 A0A182H0T4 B3NWQ4 Q9VYE9 Q8MR43 A0A1S4GMG8 A0A1B0CNY1 B4JL98 A0A1B0D0I6 B4M6P8 A0A182K314 A0A182WAW1 A0A1W4V7G2 A0A0M4ERY4 B4R4A9 A0A182R2W2 B4Q236 A0A1L8DHJ0 A0A1L8DHJ1 A0A182FLQ0 B4L223 A0A3B0JZ38 A0A1J1HY02 A0A336MHT5 B4NCI0 Q29IQ7 B3MXQ1 A0A423TVX4 A0A0L7QPS5 B7P4Y8 J9JZ53 A0A310SID5 E2AHT2 A0A154PJ62 E9IDC1 A0A2A3E7K7 A0A0K8SPS0 A0A0A9Y8V2 F7GLJ8 G9KIQ1 A0A2U9BJE1 F4WXJ8 A0A158NUY7 A0A2I4CAJ8 A0A2S2NFP4 D6X552 Q07G58 A0A443RBA7 A0A146M9R8 A0A1S3DJ30 A0A3Q2X823 A0A1A8LAH3 A0A2R5LCL1 A0A067R513 A0A1A8DAX6 A0A1A8A0N8 A0A1A8PBX4 A0A1A8JTF2 A0A1A8HIR2 A0A091LHI4 A0A151M474 A0A091J8Z8 A0A1A7WU37 H2MLH7 V9IBJ2 A0A091TJ42 A0A094L0J4 A0A3P9JIH3 A0A3P9L3L7 A0A2J7QFC9
A0A0N1IAW4 S4PMT8 A0A0K8VK64 A0A034WTL0 A0A0A1XNR8 W8C0X7 A0A0L0BU41 A0A0K8TRH0 B0WBK2 A0A1A9WZE4 A0A1A9XBP0 A0A1B0ATG2 A0A1I8N7Q3 A0A1I8QBR4 A0A182Q8Z3 A0A182NVL4 A0A182SYW4 A0A084W1A0 A0A182YI84 A0A2M4CW95 A0A182IXY3 A0A1A9ZEP2 A0A1A9VCQ1 A0A2M4CW98 A0A182H0T4 B3NWQ4 Q9VYE9 Q8MR43 A0A1S4GMG8 A0A1B0CNY1 B4JL98 A0A1B0D0I6 B4M6P8 A0A182K314 A0A182WAW1 A0A1W4V7G2 A0A0M4ERY4 B4R4A9 A0A182R2W2 B4Q236 A0A1L8DHJ0 A0A1L8DHJ1 A0A182FLQ0 B4L223 A0A3B0JZ38 A0A1J1HY02 A0A336MHT5 B4NCI0 Q29IQ7 B3MXQ1 A0A423TVX4 A0A0L7QPS5 B7P4Y8 J9JZ53 A0A310SID5 E2AHT2 A0A154PJ62 E9IDC1 A0A2A3E7K7 A0A0K8SPS0 A0A0A9Y8V2 F7GLJ8 G9KIQ1 A0A2U9BJE1 F4WXJ8 A0A158NUY7 A0A2I4CAJ8 A0A2S2NFP4 D6X552 Q07G58 A0A443RBA7 A0A146M9R8 A0A1S3DJ30 A0A3Q2X823 A0A1A8LAH3 A0A2R5LCL1 A0A067R513 A0A1A8DAX6 A0A1A8A0N8 A0A1A8PBX4 A0A1A8JTF2 A0A1A8HIR2 A0A091LHI4 A0A151M474 A0A091J8Z8 A0A1A7WU37 H2MLH7 V9IBJ2 A0A091TJ42 A0A094L0J4 A0A3P9JIH3 A0A3P9L3L7 A0A2J7QFC9
Pubmed
19121390
22118469
26354079
23622113
25348373
25830018
+ More
24495485 26108605 26369729 25315136 24438588 25244985 26483478 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 17550304 15632085 20798317 21282665 25401762 17495919 23236062 21719571 21347285 18362917 19820115 26823975 24845553 22293439 17554307
24495485 26108605 26369729 25315136 24438588 25244985 26483478 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 17550304 15632085 20798317 21282665 25401762 17495919 23236062 21719571 21347285 18362917 19820115 26823975 24845553 22293439 17554307
EMBL
BABH01003232
RSAL01000001
RVE55205.1
AGBW02009681
OWR50140.1
NWSH01000087
+ More
PCG79722.1 ODYU01001486 SOQ37751.1 KQ459596 KPI95384.1 KQ459781 KPJ19985.1 GAIX01003385 JAA89175.1 GDHF01013062 JAI39252.1 GAKP01001295 JAC57657.1 GBXI01001700 JAD12592.1 GAMC01003812 JAC02744.1 JRES01001343 KNC23531.1 GDAI01000867 JAI16736.1 DS231879 EDS42441.1 JXJN01003302 AXCN02001879 ATLV01019249 KE525266 KFB43994.1 GGFL01005434 MBW69612.1 GGFL01005435 MBW69613.1 JXUM01102040 KQ564695 KXJ71887.1 CH954180 EDV47216.1 AE014298 BT088797 AAF48249.1 ACS16655.1 AY122136 AAM52648.1 AAAB01008968 AJWK01021146 CH916370 EDW00351.1 AJVK01009940 CH940653 EDW62465.1 CP012528 ALC48965.1 CM000366 EDX17811.1 CM000162 EDX01557.1 GFDF01008155 JAV05929.1 GFDF01008156 JAV05928.1 CH933810 EDW07744.1 OUUW01000003 SPP78646.1 CVRI01000036 CRK92976.1 UFQT01001210 SSX29520.1 CH964239 EDW82539.2 CH379063 EAL32596.2 CH902630 EDV38516.1 QCYY01001101 ROT80595.1 KQ414813 KOC60559.1 ABJB010161556 ABJB010501089 DS637704 EEC01660.1 ABLF02039332 KQ767728 OAD53340.1 GL439579 EFN67011.1 KQ434936 KZC11926.1 GL762454 EFZ21394.1 KZ288344 PBC27695.1 GBRD01010710 JAG55114.1 GBHO01017624 JAG25980.1 JP016182 AES04780.1 CP026249 AWP04178.1 GL888425 EGI61068.1 ADTU01026831 ADTU01026832 GGMR01003414 MBY16033.1 KQ971382 EEZ97201.1 CR761636 CAL49463.1 NCKU01001278 RWS12552.1 GDHC01002136 JAQ16493.1 HAEF01004240 SBR41622.1 GGLE01003137 MBY07263.1 KK852747 KDR17280.1 HAEA01002960 SBQ31440.1 HADY01009718 HAEJ01000346 SBP48203.1 HAEI01001577 HAEH01006174 SBR78517.1 HAEE01003363 SBR23383.1 HAEC01014343 SBQ82560.1 KL326524 KFP56494.1 AKHW03006646 KYO19250.1 KK501569 KFP16200.1 HADW01007853 HADX01004019 SBP09253.1 JR037265 AEY57669.1 KK453579 KFQ75012.1 KL359342 KFZ64375.1 NEVH01015299 PNF27296.1
PCG79722.1 ODYU01001486 SOQ37751.1 KQ459596 KPI95384.1 KQ459781 KPJ19985.1 GAIX01003385 JAA89175.1 GDHF01013062 JAI39252.1 GAKP01001295 JAC57657.1 GBXI01001700 JAD12592.1 GAMC01003812 JAC02744.1 JRES01001343 KNC23531.1 GDAI01000867 JAI16736.1 DS231879 EDS42441.1 JXJN01003302 AXCN02001879 ATLV01019249 KE525266 KFB43994.1 GGFL01005434 MBW69612.1 GGFL01005435 MBW69613.1 JXUM01102040 KQ564695 KXJ71887.1 CH954180 EDV47216.1 AE014298 BT088797 AAF48249.1 ACS16655.1 AY122136 AAM52648.1 AAAB01008968 AJWK01021146 CH916370 EDW00351.1 AJVK01009940 CH940653 EDW62465.1 CP012528 ALC48965.1 CM000366 EDX17811.1 CM000162 EDX01557.1 GFDF01008155 JAV05929.1 GFDF01008156 JAV05928.1 CH933810 EDW07744.1 OUUW01000003 SPP78646.1 CVRI01000036 CRK92976.1 UFQT01001210 SSX29520.1 CH964239 EDW82539.2 CH379063 EAL32596.2 CH902630 EDV38516.1 QCYY01001101 ROT80595.1 KQ414813 KOC60559.1 ABJB010161556 ABJB010501089 DS637704 EEC01660.1 ABLF02039332 KQ767728 OAD53340.1 GL439579 EFN67011.1 KQ434936 KZC11926.1 GL762454 EFZ21394.1 KZ288344 PBC27695.1 GBRD01010710 JAG55114.1 GBHO01017624 JAG25980.1 JP016182 AES04780.1 CP026249 AWP04178.1 GL888425 EGI61068.1 ADTU01026831 ADTU01026832 GGMR01003414 MBY16033.1 KQ971382 EEZ97201.1 CR761636 CAL49463.1 NCKU01001278 RWS12552.1 GDHC01002136 JAQ16493.1 HAEF01004240 SBR41622.1 GGLE01003137 MBY07263.1 KK852747 KDR17280.1 HAEA01002960 SBQ31440.1 HADY01009718 HAEJ01000346 SBP48203.1 HAEI01001577 HAEH01006174 SBR78517.1 HAEE01003363 SBR23383.1 HAEC01014343 SBQ82560.1 KL326524 KFP56494.1 AKHW03006646 KYO19250.1 KK501569 KFP16200.1 HADW01007853 HADX01004019 SBP09253.1 JR037265 AEY57669.1 KK453579 KFQ75012.1 KL359342 KFZ64375.1 NEVH01015299 PNF27296.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000218220
UP000053268
UP000053240
+ More
UP000037069 UP000002320 UP000091820 UP000092443 UP000092460 UP000095301 UP000095300 UP000075886 UP000075884 UP000075901 UP000030765 UP000076408 UP000075880 UP000092445 UP000078200 UP000069940 UP000249989 UP000008711 UP000000803 UP000092461 UP000001070 UP000092462 UP000008792 UP000075881 UP000075920 UP000192221 UP000092553 UP000000304 UP000075900 UP000002282 UP000069272 UP000009192 UP000268350 UP000183832 UP000007798 UP000001819 UP000007801 UP000283509 UP000053825 UP000001555 UP000007819 UP000000311 UP000076502 UP000242457 UP000002280 UP000246464 UP000007755 UP000005205 UP000192220 UP000007266 UP000285301 UP000079169 UP000264820 UP000027135 UP000050525 UP000053119 UP000001038 UP000265200 UP000265180 UP000235965
UP000037069 UP000002320 UP000091820 UP000092443 UP000092460 UP000095301 UP000095300 UP000075886 UP000075884 UP000075901 UP000030765 UP000076408 UP000075880 UP000092445 UP000078200 UP000069940 UP000249989 UP000008711 UP000000803 UP000092461 UP000001070 UP000092462 UP000008792 UP000075881 UP000075920 UP000192221 UP000092553 UP000000304 UP000075900 UP000002282 UP000069272 UP000009192 UP000268350 UP000183832 UP000007798 UP000001819 UP000007801 UP000283509 UP000053825 UP000001555 UP000007819 UP000000311 UP000076502 UP000242457 UP000002280 UP000246464 UP000007755 UP000005205 UP000192220 UP000007266 UP000285301 UP000079169 UP000264820 UP000027135 UP000050525 UP000053119 UP000001038 UP000265200 UP000265180 UP000235965
Interpro
ProteinModelPortal
H9JEJ8
A0A3S2LJF9
A0A212F8U1
A0A2A4K7U1
A0A2H1VAC8
A0A194PRK7
+ More
A0A0N1IAW4 S4PMT8 A0A0K8VK64 A0A034WTL0 A0A0A1XNR8 W8C0X7 A0A0L0BU41 A0A0K8TRH0 B0WBK2 A0A1A9WZE4 A0A1A9XBP0 A0A1B0ATG2 A0A1I8N7Q3 A0A1I8QBR4 A0A182Q8Z3 A0A182NVL4 A0A182SYW4 A0A084W1A0 A0A182YI84 A0A2M4CW95 A0A182IXY3 A0A1A9ZEP2 A0A1A9VCQ1 A0A2M4CW98 A0A182H0T4 B3NWQ4 Q9VYE9 Q8MR43 A0A1S4GMG8 A0A1B0CNY1 B4JL98 A0A1B0D0I6 B4M6P8 A0A182K314 A0A182WAW1 A0A1W4V7G2 A0A0M4ERY4 B4R4A9 A0A182R2W2 B4Q236 A0A1L8DHJ0 A0A1L8DHJ1 A0A182FLQ0 B4L223 A0A3B0JZ38 A0A1J1HY02 A0A336MHT5 B4NCI0 Q29IQ7 B3MXQ1 A0A423TVX4 A0A0L7QPS5 B7P4Y8 J9JZ53 A0A310SID5 E2AHT2 A0A154PJ62 E9IDC1 A0A2A3E7K7 A0A0K8SPS0 A0A0A9Y8V2 F7GLJ8 G9KIQ1 A0A2U9BJE1 F4WXJ8 A0A158NUY7 A0A2I4CAJ8 A0A2S2NFP4 D6X552 Q07G58 A0A443RBA7 A0A146M9R8 A0A1S3DJ30 A0A3Q2X823 A0A1A8LAH3 A0A2R5LCL1 A0A067R513 A0A1A8DAX6 A0A1A8A0N8 A0A1A8PBX4 A0A1A8JTF2 A0A1A8HIR2 A0A091LHI4 A0A151M474 A0A091J8Z8 A0A1A7WU37 H2MLH7 V9IBJ2 A0A091TJ42 A0A094L0J4 A0A3P9JIH3 A0A3P9L3L7 A0A2J7QFC9
A0A0N1IAW4 S4PMT8 A0A0K8VK64 A0A034WTL0 A0A0A1XNR8 W8C0X7 A0A0L0BU41 A0A0K8TRH0 B0WBK2 A0A1A9WZE4 A0A1A9XBP0 A0A1B0ATG2 A0A1I8N7Q3 A0A1I8QBR4 A0A182Q8Z3 A0A182NVL4 A0A182SYW4 A0A084W1A0 A0A182YI84 A0A2M4CW95 A0A182IXY3 A0A1A9ZEP2 A0A1A9VCQ1 A0A2M4CW98 A0A182H0T4 B3NWQ4 Q9VYE9 Q8MR43 A0A1S4GMG8 A0A1B0CNY1 B4JL98 A0A1B0D0I6 B4M6P8 A0A182K314 A0A182WAW1 A0A1W4V7G2 A0A0M4ERY4 B4R4A9 A0A182R2W2 B4Q236 A0A1L8DHJ0 A0A1L8DHJ1 A0A182FLQ0 B4L223 A0A3B0JZ38 A0A1J1HY02 A0A336MHT5 B4NCI0 Q29IQ7 B3MXQ1 A0A423TVX4 A0A0L7QPS5 B7P4Y8 J9JZ53 A0A310SID5 E2AHT2 A0A154PJ62 E9IDC1 A0A2A3E7K7 A0A0K8SPS0 A0A0A9Y8V2 F7GLJ8 G9KIQ1 A0A2U9BJE1 F4WXJ8 A0A158NUY7 A0A2I4CAJ8 A0A2S2NFP4 D6X552 Q07G58 A0A443RBA7 A0A146M9R8 A0A1S3DJ30 A0A3Q2X823 A0A1A8LAH3 A0A2R5LCL1 A0A067R513 A0A1A8DAX6 A0A1A8A0N8 A0A1A8PBX4 A0A1A8JTF2 A0A1A8HIR2 A0A091LHI4 A0A151M474 A0A091J8Z8 A0A1A7WU37 H2MLH7 V9IBJ2 A0A091TJ42 A0A094L0J4 A0A3P9JIH3 A0A3P9L3L7 A0A2J7QFC9
Ontologies
PANTHER
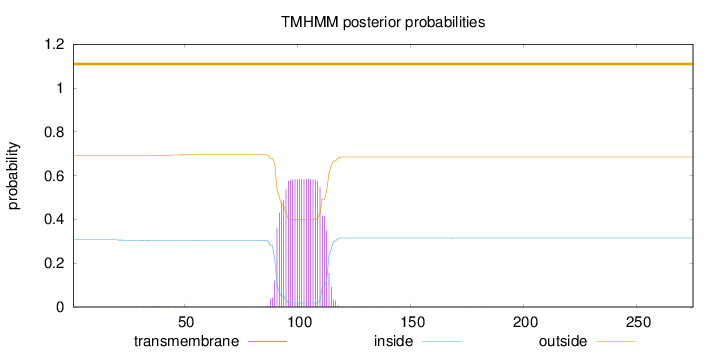
Topology
Length:
275
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.77787
Exp number, first 60 AAs:
0.07712
Total prob of N-in:
0.30664
outside
1 - 275
Population Genetic Test Statistics
Pi
237.000493
Theta
220.676864
Tajima's D
-1.268615
CLR
0.847331
CSRT
0.0918954052297385
Interpretation
Uncertain
