Gene
KWMTBOMO08648 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007948
Annotation
PREDICTED:_spectrin_alpha_chain_isoform_X1_[Bombyx_mori]
Full name
Spectrin alpha chain
Location in the cell
Cytoplasmic Reliability : 1.878 Nuclear Reliability : 1.239
Sequence
CDS
ATGGAGCAGATCCCACCACCGAAAGAGGTGAAGATCCTCGAAACAGCAGAGGACATTCAGGAGCGTCGTGAGGAGGTTTTGAACCGCTATGAAGAGTTTAAGCAGGAAGCCCGTGCCAAACGTGAGAAACTCGAGGACTCCCGTCGCTTCCAGTACTTCAAGCGTGATGCTGATGAACTCGAGTCGTGGATTCAGGAGAAGCTTCAGGCCGCCAGCGATGAGAGCTACAAAGATCCCACCAACCTTCAGGCGAAAATCCAGAAACATCAAGCATTCGAAGCGGAGGTAGCTGCGCACTCCAATGCTATTGTAGTTTTGGACAACACCGGGTCAGAGATGATAGCTGCGGGACACTTCGCTTCGGAGACCATCCGTCGGCGCTTGGACGAACTACACCGTCTCTGGGAACTGTTGCTGTCCCGCCTCGCCGAGAAGGGCATGAAGCTGCAGCAGGCGCTTGTACTGGTGCAGTTTCTGAGGCACTGCGATGAAGTCATGTTCTGGATTCATGACAAGGAAACGTTTGTGTGCGCGGATGAGTTCGGTTCTGACCTGGAGCATGTTGAGGTGCTTCAGCGCAAGTTCGACGAGTTCCAGAAGGACATGGCCGCCCAGGAGTACCGGGTCACTGAAGTCAACCAGCTGGCCGAGAGACTCGTACTCGAAGGACACCCTGAAAGGGAAACCATCGTCAAGAGAAAAGACGAGTTGAACGAAGCCTGGCAGCGCCTGCGTCAAATGGCACTCATGCGTCAAGAACGTTTGTTCGGAGCCCACGAGATCCAGCGCTTCAATCGCGATGCCGACGAGACCATCGCCTGGATCTCGGAGAAGGATGCCGTGCTCGGATCCGACGACTACGGGCGCGACCTGGCTACCGTGCAGACGCTACAGCGCAAGCACGAAGGAGTAGAGCGCGATCTGGCCGCGTTAGAGGACAAGGTGTCGGTGCTGGAAGGAGAGGCCGTCCGCCTGGGGGCCATCCACGGCGCGCACGCCGACGCCATCAGCGCCAAGCGCGACGAGGCGCACGCGGCCTGGCAGCGCCTGGTGCAGCGCGCCGCGCACCGCCGGCAGCGCCTCGAGGCCGCGCTCGCGCTGCACCGCTTCCTGGCCGACTACCGCGACCTGCTCTCCTGGCTCGCAGACATGCGGGCGCTCATCGCCGCCGACGACCTCGCCAAGGACGTGCCCGGGGCCGAGGCTCTGCTCGAGAGGCACCAGGAGCACAAGGGCGAGATGGACGCGCGCGCGGACGTGATCGGCGCGTGTGCCGTGGCGGGGCAGGCGCTGGTGGCCAGCGAGCACCACGCGGCGGCCGACGTGTCCGCGGCGCTGCACACGCTGGAGCGGGAGGCCGCAGCGCTGCAGCAGCTGTGGGAGCAGCGCCGCGTGCTCTACCTGCAGTGCATGGACCTGCAACTCTTCTACCGCGACACTGAACAAGCCGACGCCTGGATGCACAAACAAGAGGCTTTCCTTGCGAACGAAGATGTAGGAGACTCTTTAGACTCCGTGGAAGCTTTGCTGAAGAAACACGAAGACTTTGAAAAGTCACTCGCCGCGCAAGAAGAGAAGATCAAGGCCCTAGACGAGTTCGCTACCAAACTTATTGAGGGCAACCATTACGCCGCCGACGATGTCGCACAAAGACGAGAGATGTTGTTGGAGCGTCGTGCGGCTTTACTAGAGAAGTCGAACCAGCGCCGCGCTCTCCTCGAAGACGCCTACAAGTATCAACAGTTCGAACGCGATTGTGATGAAACTAAAGGCTGGGTAAACGAGAAGCTGAAGTTCGCCACCGACGACTCGTACTTGGATCCGACCAACCTGAATGGCAAGGTGCAGAAACATCAGAACTTCGAACAGGAGCTGCAGGCCAACAAGCCTCGCGTCGACGAGATAACGGCCGTCGGCAAGCGTCTGCTTGAGCTCGAACATTTTGCTCAACCTCAAATATCGTCTCGCGTGGAAGAGCTGGGCGGAATATGGGAGCGACTGGTGCAGTCTTCGGAGCTGAAGGGAAACAAGTTGCAGGAAGCGGCTCAACAACAGCAGTACAATCGGGCCGTCGAGGACATTGAACTTTGGCTCTCGGAAGTTGAAGGACAGCTGCTCAGCGAGGACTACGGGAAGGATCTGACCAGCGTGCAGAATCTCCAGAAGAAGCACGCGCTGTTGGAGGCCGACGTGAGCTCGCACGCCGAACGCATCGACGCCATACGGAACCAGGCCGAGCAGTTCATAGAGAAGGGACACTTCGACGCGGATAACATCAAGGCTAAGCGGGACGCGTTGGTGGGCCGATACGCGGCTCTGGATGCGCCCATGGCGGTGCGCAAGCGGAGGCTGCTGGACTCGCTGCAGGCGCAGCAGCTGTTCCGCGACCTGGACGACGAGGCCGCCTGGATACGGGAGAAGGAGCCCGTAATTGCGTCCACGAACAGAGGTCGCGACCTGATCGGCGTCCAGAACCTTATGAAGAAGCACCAGGCCGTGCTGGGCGAGATGGCGCAGCACGAGGCGCGCGTGGAGGCGGTGCGGGCGGCCGGGGCTGCGCTGCGGGACGCCGGACACTTCGCCGCCGCCGACATCGCCGCCAGACTCACGCAGCTCCGCGACAACTGGACACAGCTGCAGGAGAAGGCGCTGCAGCGTAAACAAGACCTCGAAGACTCTCTTCAAGCTCAGCAGTACTTTGCTGATGCCAACGAAGCTGAATCTTGGATGAGAGAGAAGGAGCCCATCGCCTGCACCCAGGACTACGGAAAGGACGAGGACTCCTCTGAGGCGCTGCTAAAGAAACACGAGGCCTTGCTGTCTGACTTGGAAGCTTTCGGGAATACCATCAAGTCGCTGAGGGAACAGGCCAATTCATGCAGACAGCAGGAGTCGCCCGTGGTGGACGAGTCGGGCAAGGAGTGCGTGGCGGCGCTGTACGACTACGCGGAGAAGTCACCGCGCGAGGTCTCCATGAAGCGCGGCGACGTGCTCACGCTGCTCAACTCCAACAACAAGGACTGGTGGAAGGTGGAAGTAAACGACCGCCAAGGCTTCGTCCCGGCCGCATACGTCAAGAAGATCGACGCCGGACTGTCGTCCTCCCAGCAGAACCTCGCCGACTCCAACTCCATCGCCGCCAGACAGAATCAGATCGAGTCGCAGTATGACAACCTGCTGGGGCTGGCGCGCGAGCGGCAGAACAAACTCAACGAGACCGTCAAGGCCTACGTGCTGGTGCGGGAGGCGGCCGAGCTCGCCACCTGGATCAAGGACAAGGAGATGCACGCTCAAGTTCAAGACGTGGGCGAAGATCTGGAGCAGGTCGAGGTGATGCAGAAGAAGTTCGACGACTTCCAGAACGACCTGCGCGCCAACGAGGTGCGGCTCGCCGAGATGAACGAGATCGCCGTGCAGCTGATGACCGTCGGACAGACCGAGGCAGCCCTGAAGATCAAGACTCAGATGCAGGAGCTGAACTCGAAGTGGAGCTCGCTGCAGCAGCTGACGGCGGAGCGCGCGGCGCAGCTGGGCTCGGCGCACGAGGTGCAGCGCTTCCACCGCGACGTCGACGAGACCAAGGACTGGATCGCGGAGAAGGACGCCGCGCTCGCCTCCGACGACCTGGGCCGCGACCTGCGCTCCGTGCAGACCCTGCAGCGCAAGCATGAAGGCCTAGAGCGCGATTTAGCAGCCCTCGGCGACAAGATCCGCCAACTGGACGACACCGCCAACCGGCTGATGTCCACGCACGGGGACTCCGCCGACGCTACCTACAGCAAGCAGATAGAGATCAACGAAGCCTGGCAGCAACTGCAGGCGCGCGCCAATGCCAGGAAGGAGAAGCTTCTGGACTCCTACGACCTGCAGAGGTTCTTGTCGGACTACCGCGACTTGATGGCGTGGATCAACTCGATGATGGCGCTGGTGAGCTCGGACGAACTCGCCAATGACGTCACCGGCGCTGAGGCCCTGCTGGAAAGACATCAGGAGCACCGTACGGAGATGGACGCTCGCGCGGGCACGTTCCAGGCTCTGGAACTGTTCGGCCAGCAGCTGCTGCAGGGCGGCCACTACGCCAGCGTCGACATACAGGAGAAGCTCAACAACATGAGCGACGCCAGACAGGAACTCGAGAAAGCTTGGGTGAACCGACGCATGAAGTTAGACCAGAACTTGGAGCTGCAGCTGTTCTACCGTGACTGCGAGCAGGCGGAGGGCTGGATGGCCGCTCGCGAGGCCTTCCTGGCGCCCGCGGACCACGCCGACTCCGCCGACCAGCAGCAGCCGGACAACGTCGAGCAACTCATCAAGAAACACGAAGACTTCGACAAGGCCATCAATGCTCACGAGGAGAAGATTGCCCAGCTGCAGACGTTGGCCGACCAGCTGATCGCTTCCGACCACTACGCGACCGACGACATCGACGACAAGCGCAAGCAGGTGCTCGACCGCTGGAGGCACCTCAAGGAGGCACTCATAGAGAAACGCTCCAGATTGGGTGATGAACAAACTCTGCAACAATTCTCTCGCGACGCTGACGAGATGGAGAATTGGATCATGGAGAAGATGCAGTTAGCTACTGAAGAGAGTTACAAGGACCCAACCAATATTCAGTCCAAACATCAGAAGCACCAGGCGTTCGAAGCAGAACTGGCGGCCAACGCCGAGCGCATCCAGTCCGTGCTCGCCATGGGAGGCAACCTCGTGCAGCGCGGACAGTGCAGCGGCAGCGAGGATGCCGTGCAGGCTCGTCTCGCTTCCATCGCTGACCAATGGGAGTACCTCACTCAGAAGACTACAGAGAAGTCTCTGAAATTGAAAGAAGCGAACAAGCAGCGGACTTACATTGCTGCCGTTAAAGATCTGGACTTCTGGCTCGGTGAGGTCGAGAGCTTGTTGACCTCAGAAGACTCCGGCAAGGACCTGGCTTCTGTTCAGAACTTAATGAAGAAACACCAGCTCGTGGAAGCTGATATCCAAGCTCACGAGGACAGAATCAATGACATGAACGGGCAGGCGGACGCGCTGGTGGAGAGCGGGCAGTTCGACAGCGCCGGCATCGGCTCGCGCCGCGACGCCATCAACGAGCGCTTCGAGCGCGTGCGCAACCTCGCCGCGCACCGCCGCGCCAGACTGCACGAGGCCAACACGCTGCACCAGTTCTTCAGGGACATCGCCGACGAGGAGTCCTGGATCAAGGAGAAAAAACTTTTGGTCGCTTCGGATGACTACGGCCGCGACTTGACCGGCGTCCAGAACTTACTGAAGAAGCACAAGCGTCTCGAAGCGGAACTTGCGAGTCACGAGCCCGCCATACAAGCCGTGCAGGAGGCCGGCGAGAAGCTGATGGACGTGTCCAACCTTGGCGTGCTCGAGATCGAACAGCGGCTGAAAGCTCTGGCCTTGGCCTGGGATGAACTCCAAGCGCTGGCTGCCGAGCGAGGGGCCAAGCTGCAGCAGTCGCTCGCCTACCAACAGTTCCTTGCCAAGGTTGACGAAGAGGAGGCCTGGATCAGCGAGAAGCAGCAGCTGGTGGCGGTGGGCGAGTGCGGAGACAGCATGGCGGCGGTGCAGGGCCTGCTGAAGAAGCACGAGGCGCTGGAGGCGGAGCTGGCGGCGCGCGGCGAGCGGGTGCGCGAGCTGGCGGCGGCGGGCGACGCGCTGCTGGCGGCCGGCAACCTGCACCACGACGCGCTGGCGCACCGCATCCGCGCCCTGCAGGCCAAACTGGAAAAGCTCACGGCATTGGCGGCTCGTCGCAAGGTAGCCCTTGTAGACAACTCCCTCTATCTGCAGCTACTGTGGAAGGCCGACGTGGTGGAATCCTGGATCGCAGACAAGGAGACACACGTGCGATCTGATGAGTTCGGTCGAGATCTGTCCACTGTGCAGACATTGCTCACCAAGCAAGACACTTTCGATGCTGGTCTGGCGGCGTTCGAGCACGAAGGGATCCAGAACATCACGTCTCTGAAAGAGCAGCTGGTGGCGGCGGGGCACGAGCAGACGCCCGCCATCGTGAAGCGGCACGGTGACGTCATCGCGCGCTGGCAGCGACTGCTCTCCGACTCCGCCACCAGGAAGCAACGGCTGCTGCAGCTGCAGGACCAGTTCCGACAGATCGAGGAACTCTACCTCACCTTCGCCAAAAAGGCTTCTGCATTCAACTCGTGGTTCGAGAACGCGGAGGAGGACCTGACGGACCCCGTGCGCTGCAACTCCATCGAGGAGATACGCGCGCTCAAGGACGCGCACGCGCAGTTCCAGGCGAGTGACCCACGCATATTCCAACTGTATCCCGCCTCGGAGCGAGACGTGGAGTTGTCGAAGGAAGCTCAACGGCAGGAGGAGAACGACAAGCTGCGGAAGGAGTTCGCCAAGCACGCCAACACCTTCCACCAGTGGCTCACCGAGACTCGGTACCGGTTGCTAGGCTGGGACGGCACATCAATGATGGAGGGCACGGGTTCCCTGGAACAACAGTTGGCGACGCTGCGCCAACGAGCGTCCGAAGTACGCGCCCGCAGAGCTGACCTGAGACGCCTTGAAGAGTTAGGCGCCGCCCTGGAGGAGCATCTGATCCTGGACAACCGCTACACGGAGCACAGCACCGTGGGACTAGCGCAGCAGTGGGACCAGCTGGACCAGCTCTCTATGCGCATGCAGCACAACCTCGAGCAGCAGATACAAGCCAGGAACCATTCCGGTGTAAGCGAAGACGCCCTGAAGGAGTTCTCGATGATGTTCAAGCACTTCGACAAGGATCGATCTGGACGCCTCAACCACCACGAGTTCAAGTCGTGTCTGCGAGCCCTCGGGTACGACCTGCCCATGGTTGAAGAGGGACAACCCGATCCTGAGTTTGAATCTATACTCAGTATCGTAGATCCGAACCGCGACGGCCAAGTATCCCTTCAAGAGTACATGGCCTTCATGATCAGCAAGGAGACCGAGAACGTGCACTCCTCCGAGGAGATCGAGAACGCGTTCCGCGCCATCACCGCGCACCAGGACCGGCCCTACGTCACCAAGGAAGAACTGTACGCGAACCTCACGAAGGAGATGGCGGACTACTGCGTGGCGCGCATGAAGCCCTACGTGGAGCCGAAGACGGAGCGCCCCATACCCAACGCGCTCGACTACATCGACTTCACACACACTCTGTTCCAAAACTAA
Protein
MEQIPPPKEVKILETAEDIQERREEVLNRYEEFKQEARAKREKLEDSRRFQYFKRDADELESWIQEKLQAASDESYKDPTNLQAKIQKHQAFEAEVAAHSNAIVVLDNTGSEMIAAGHFASETIRRRLDELHRLWELLLSRLAEKGMKLQQALVLVQFLRHCDEVMFWIHDKETFVCADEFGSDLEHVEVLQRKFDEFQKDMAAQEYRVTEVNQLAERLVLEGHPERETIVKRKDELNEAWQRLRQMALMRQERLFGAHEIQRFNRDADETIAWISEKDAVLGSDDYGRDLATVQTLQRKHEGVERDLAALEDKVSVLEGEAVRLGAIHGAHADAISAKRDEAHAAWQRLVQRAAHRRQRLEAALALHRFLADYRDLLSWLADMRALIAADDLAKDVPGAEALLERHQEHKGEMDARADVIGACAVAGQALVASEHHAAADVSAALHTLEREAAALQQLWEQRRVLYLQCMDLQLFYRDTEQADAWMHKQEAFLANEDVGDSLDSVEALLKKHEDFEKSLAAQEEKIKALDEFATKLIEGNHYAADDVAQRREMLLERRAALLEKSNQRRALLEDAYKYQQFERDCDETKGWVNEKLKFATDDSYLDPTNLNGKVQKHQNFEQELQANKPRVDEITAVGKRLLELEHFAQPQISSRVEELGGIWERLVQSSELKGNKLQEAAQQQQYNRAVEDIELWLSEVEGQLLSEDYGKDLTSVQNLQKKHALLEADVSSHAERIDAIRNQAEQFIEKGHFDADNIKAKRDALVGRYAALDAPMAVRKRRLLDSLQAQQLFRDLDDEAAWIREKEPVIASTNRGRDLIGVQNLMKKHQAVLGEMAQHEARVEAVRAAGAALRDAGHFAAADIAARLTQLRDNWTQLQEKALQRKQDLEDSLQAQQYFADANEAESWMREKEPIACTQDYGKDEDSSEALLKKHEALLSDLEAFGNTIKSLREQANSCRQQESPVVDESGKECVAALYDYAEKSPREVSMKRGDVLTLLNSNNKDWWKVEVNDRQGFVPAAYVKKIDAGLSSSQQNLADSNSIAARQNQIESQYDNLLGLARERQNKLNETVKAYVLVREAAELATWIKDKEMHAQVQDVGEDLEQVEVMQKKFDDFQNDLRANEVRLAEMNEIAVQLMTVGQTEAALKIKTQMQELNSKWSSLQQLTAERAAQLGSAHEVQRFHRDVDETKDWIAEKDAALASDDLGRDLRSVQTLQRKHEGLERDLAALGDKIRQLDDTANRLMSTHGDSADATYSKQIEINEAWQQLQARANARKEKLLDSYDLQRFLSDYRDLMAWINSMMALVSSDELANDVTGAEALLERHQEHRTEMDARAGTFQALELFGQQLLQGGHYASVDIQEKLNNMSDARQELEKAWVNRRMKLDQNLELQLFYRDCEQAEGWMAAREAFLAPADHADSADQQQPDNVEQLIKKHEDFDKAINAHEEKIAQLQTLADQLIASDHYATDDIDDKRKQVLDRWRHLKEALIEKRSRLGDEQTLQQFSRDADEMENWIMEKMQLATEESYKDPTNIQSKHQKHQAFEAELAANAERIQSVLAMGGNLVQRGQCSGSEDAVQARLASIADQWEYLTQKTTEKSLKLKEANKQRTYIAAVKDLDFWLGEVESLLTSEDSGKDLASVQNLMKKHQLVEADIQAHEDRINDMNGQADALVESGQFDSAGIGSRRDAINERFERVRNLAAHRRARLHEANTLHQFFRDIADEESWIKEKKLLVASDDYGRDLTGVQNLLKKHKRLEAELASHEPAIQAVQEAGEKLMDVSNLGVLEIEQRLKALALAWDELQALAAERGAKLQQSLAYQQFLAKVDEEEAWISEKQQLVAVGECGDSMAAVQGLLKKHEALEAELAARGERVRELAAAGDALLAAGNLHHDALAHRIRALQAKLEKLTALAARRKVALVDNSLYLQLLWKADVVESWIADKETHVRSDEFGRDLSTVQTLLTKQDTFDAGLAAFEHEGIQNITSLKEQLVAAGHEQTPAIVKRHGDVIARWQRLLSDSATRKQRLLQLQDQFRQIEELYLTFAKKASAFNSWFENAEEDLTDPVRCNSIEEIRALKDAHAQFQASDPRIFQLYPASERDVELSKEAQRQEENDKLRKEFAKHANTFHQWLTETRYRLLGWDGTSMMEGTGSLEQQLATLRQRASEVRARRADLRRLEELGAALEEHLILDNRYTEHSTVGLAQQWDQLDQLSMRMQHNLEQQIQARNHSGVSEDALKEFSMMFKHFDKDRSGRLNHHEFKSCLRALGYDLPMVEEGQPDPEFESILSIVDPNRDGQVSLQEYMAFMISKETENVHSSEEIENAFRAITAHQDRPYVTKEELYANLTKEMADYCVARMKPYVEPKTERPIPNALDYIDFTHTLFQN
Summary
Description
Spectrin is the major constituent of the cytoskeletal network underlying the erythrocyte plasma membrane. It associates with band 4.1 and actin to form the cytoskeletal superstructure of the erythrocyte plasma membrane. Essential for larval survival and development. Stabilizes cell to cell interactions that are critical for the maintenance of cell shape and subcellular organization within embryonic tissues. Lva and spectrin may form a Golgi-based scaffold that mediates interaction of Golgi bodies with microtubules and facilitates Golgi-derived membrane secretion required for the formation of furrows during cellularization.
Subunit
Native spectrin molecule is a tetramer composed of two antiparallel heterodimers joined head to head so that each end of the native molecule includes the C-terminus of the alpha subunit and the N-terminus of the beta subunit. Interacts with calmodulin in a calcium-dependent manner, interacts with F-actin and also interacts with Lva. Interacts with Ten-m.
Miscellaneous
Its transcript shares the first untranslated exon with the dlt transcript, suggesting a common regulation.
Similarity
Belongs to the spectrin family.
Keywords
3D-structure
Actin capping
Actin-binding
Calcium
Calmodulin-binding
Cell shape
Complete proteome
Cytoplasm
Cytoskeleton
Golgi apparatus
Metal-binding
Phosphoprotein
Reference proteome
Repeat
SH3 domain
Feature
chain Spectrin alpha chain
Uniprot
A0A1E1WR33
A0A2H1VAD9
A0A173FDD0
S4PH37
A0A212FB96
A0A437BXV0
+ More
A0A2J7QVA1 A0A2J7QV98 A0A067RUI8 A0A069DXZ1 A0A1S3DD98 A0A224X4N5 A0A0V0G435 A0A2J7QVA8 A0A0K8SCT5 E0VV65 A0A0P4VTQ5 V9IJN9 A0A0K8TCJ0 A0A158NIL4 T1HPS9 A0A088ACE3 E9I8D6 A0A0C9R692 A0A151J4T0 A0A151XDV2 A0A195BCT0 A0A158NIL3 A0A2A3EFW3 A0A026W3N3 A0A151IP53 E2AX44 A0A195FLX6 A0A1W4XPA6 A0A411G645 A0A154P796 E2BFC6 K7J3F4 A0A1Y1LRY4 A0A0R1DUS3 A0A0Q5U4D6 D6W8J4 M9PDQ0 A0A0Q9XL73 A0A0P8ZSY5 A0A1W4W6C4 W8ANY7 F4WS96 A0A0R3P5Q6 A0A0Q5U2Q0 A0A0Q9XBE9 P13395 A0A232FF24 A0A0R1DZX8 A0A0Q5U8T3 A0A0R1DUG6 A0A1W4VTS2 A0A034VFQ9 A0A0Q9XAW4 A0A1I8ME62 A0A0P8Y852 W8B358 A0A0P9C1U7 T1PNU9 M9PGV6 A0A1Y1LU64 A0A1I8QDV7 A0A0Q9W9D3 A0A0L0CNI9 Q29EZ8 B4MXT6 A0A1L8E5B8 A0A0A1WZX6 A0A3B0KFZ2 B4KXU7 A0A1A9V2Y6 A0A1Q3FEY7 B4HW03 A0A1B0B477 A0A0R3P5T5 A0A2S2PAP5 A0A2H8TYT6 A0A1A9W748 A0A1I8ME59 B3M7A4 Q16EQ1 T1PDG2 A0A1I8ME61 A0A0M4F0U0 A0A0K8UAC6 A0A034VI07 M9PBI5 B4PDU1 B3NEV4 A0A1I8QDR3 A0A182H9F8 A0A1I8QDP4 A0A2S2QHA8 B4MGM3 A0A0A1XG25
A0A2J7QVA1 A0A2J7QV98 A0A067RUI8 A0A069DXZ1 A0A1S3DD98 A0A224X4N5 A0A0V0G435 A0A2J7QVA8 A0A0K8SCT5 E0VV65 A0A0P4VTQ5 V9IJN9 A0A0K8TCJ0 A0A158NIL4 T1HPS9 A0A088ACE3 E9I8D6 A0A0C9R692 A0A151J4T0 A0A151XDV2 A0A195BCT0 A0A158NIL3 A0A2A3EFW3 A0A026W3N3 A0A151IP53 E2AX44 A0A195FLX6 A0A1W4XPA6 A0A411G645 A0A154P796 E2BFC6 K7J3F4 A0A1Y1LRY4 A0A0R1DUS3 A0A0Q5U4D6 D6W8J4 M9PDQ0 A0A0Q9XL73 A0A0P8ZSY5 A0A1W4W6C4 W8ANY7 F4WS96 A0A0R3P5Q6 A0A0Q5U2Q0 A0A0Q9XBE9 P13395 A0A232FF24 A0A0R1DZX8 A0A0Q5U8T3 A0A0R1DUG6 A0A1W4VTS2 A0A034VFQ9 A0A0Q9XAW4 A0A1I8ME62 A0A0P8Y852 W8B358 A0A0P9C1U7 T1PNU9 M9PGV6 A0A1Y1LU64 A0A1I8QDV7 A0A0Q9W9D3 A0A0L0CNI9 Q29EZ8 B4MXT6 A0A1L8E5B8 A0A0A1WZX6 A0A3B0KFZ2 B4KXU7 A0A1A9V2Y6 A0A1Q3FEY7 B4HW03 A0A1B0B477 A0A0R3P5T5 A0A2S2PAP5 A0A2H8TYT6 A0A1A9W748 A0A1I8ME59 B3M7A4 Q16EQ1 T1PDG2 A0A1I8ME61 A0A0M4F0U0 A0A0K8UAC6 A0A034VI07 M9PBI5 B4PDU1 B3NEV4 A0A1I8QDR3 A0A182H9F8 A0A1I8QDP4 A0A2S2QHA8 B4MGM3 A0A0A1XG25
Pubmed
27251284
23622113
22118469
24845553
26334808
20566863
+ More
27129103 26823975 21347285 21282665 24508170 30249741 20798317 20075255 28004739 17994087 17550304 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 21719571 15632085 2808524 8276898 12537569 3680372 11076973 2497103 14667407 17372656 18327897 22426000 8266097 28648823 18057021 25348373 25315136 26108605 25830018 17510324 26483478
27129103 26823975 21347285 21282665 24508170 30249741 20798317 20075255 28004739 17994087 17550304 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 21719571 15632085 2808524 8276898 12537569 3680372 11076973 2497103 14667407 17372656 18327897 22426000 8266097 28648823 18057021 25348373 25315136 26108605 25830018 17510324 26483478
EMBL
GDQN01001748
JAT89306.1
ODYU01001486
SOQ37756.1
KT182638
ANG83465.1
+ More
GAIX01003407 JAA89153.1 AGBW02009363 OWR50993.1 RSAL01000001 RVE55210.1 NEVH01010478 PNF32510.1 PNF32509.1 KK852460 KDR23504.1 GBGD01000026 JAC88863.1 GFTR01008961 JAW07465.1 GECL01003684 JAP02440.1 PNF32511.1 GBRD01014730 JAG51096.1 DS235803 EEB17271.1 GDKW01000580 JAI56015.1 JR050446 AEY61280.1 GBRD01002548 GDHC01007121 JAG63273.1 JAQ11508.1 ADTU01016945 ADTU01016946 ACPB03012376 ACPB03012377 GL761538 EFZ23210.1 GBYB01011829 JAG81596.1 KQ980088 KYN17783.1 KQ982254 KYQ58537.1 KQ976513 KYM82376.1 KZ288256 PBC30627.1 KK107503 QOIP01000006 EZA49654.1 RLU21479.1 KQ976891 KYN07257.1 GL443520 EFN61994.1 KQ981430 KYN41700.1 MH365539 QBB01348.1 KQ434831 KZC07799.1 GL448009 EFN85586.1 GEZM01048770 JAV76419.1 CM000159 KRK00797.1 CH954178 KQS43116.1 KQ971312 EEZ99233.1 AE014296 AGB93983.1 CH933809 KRG05646.1 CH902618 KPU77622.1 GAMC01018838 JAB87717.1 GL888307 EGI62932.1 CH379070 KRT08481.1 KQS43115.1 KRG05645.1 M26400 BT023889 S67762 S67765 AY069741 NNAY01000310 OXU29302.1 KRK00794.1 KQS43114.1 KQS43117.1 KRK00795.1 KRK00796.1 GAKP01016822 GAKP01016821 JAC42131.1 KRG05647.1 KPU77623.1 GAMC01018839 JAB87716.1 KPU77621.1 KA649638 AFP64267.1 AGB93981.1 GEZM01048771 JAV76418.1 CH940682 KRF77567.1 JRES01000247 KNC33004.1 EAL29911.1 CH963876 EDW76855.1 GFDF01000153 JAV13931.1 GBXI01010096 JAD04196.1 OUUW01000009 SPP85249.1 EDW17619.1 GFDL01008932 JAV26113.1 CH480817 EDW50118.1 JXJN01008189 KRT08482.1 GGMR01013845 MBY26464.1 GFXV01007639 MBW19444.1 EDV38765.1 CH478619 EAT32709.1 KA646827 AFP61456.1 CP012525 ALC44812.1 GDHF01028761 GDHF01025122 GDHF01013738 GDHF01013593 GDHF01009620 GDHF01002806 JAI23553.1 JAI27192.1 JAI38576.1 JAI38721.1 JAI42694.1 JAI49508.1 GAKP01016823 GAKP01016820 JAC42129.1 AGB93982.1 EDW92906.1 EDV50227.1 JXUM01030164 JXUM01030165 GGMS01007914 MBY77117.1 EDW57089.1 GBXI01003963 JAD10329.1
GAIX01003407 JAA89153.1 AGBW02009363 OWR50993.1 RSAL01000001 RVE55210.1 NEVH01010478 PNF32510.1 PNF32509.1 KK852460 KDR23504.1 GBGD01000026 JAC88863.1 GFTR01008961 JAW07465.1 GECL01003684 JAP02440.1 PNF32511.1 GBRD01014730 JAG51096.1 DS235803 EEB17271.1 GDKW01000580 JAI56015.1 JR050446 AEY61280.1 GBRD01002548 GDHC01007121 JAG63273.1 JAQ11508.1 ADTU01016945 ADTU01016946 ACPB03012376 ACPB03012377 GL761538 EFZ23210.1 GBYB01011829 JAG81596.1 KQ980088 KYN17783.1 KQ982254 KYQ58537.1 KQ976513 KYM82376.1 KZ288256 PBC30627.1 KK107503 QOIP01000006 EZA49654.1 RLU21479.1 KQ976891 KYN07257.1 GL443520 EFN61994.1 KQ981430 KYN41700.1 MH365539 QBB01348.1 KQ434831 KZC07799.1 GL448009 EFN85586.1 GEZM01048770 JAV76419.1 CM000159 KRK00797.1 CH954178 KQS43116.1 KQ971312 EEZ99233.1 AE014296 AGB93983.1 CH933809 KRG05646.1 CH902618 KPU77622.1 GAMC01018838 JAB87717.1 GL888307 EGI62932.1 CH379070 KRT08481.1 KQS43115.1 KRG05645.1 M26400 BT023889 S67762 S67765 AY069741 NNAY01000310 OXU29302.1 KRK00794.1 KQS43114.1 KQS43117.1 KRK00795.1 KRK00796.1 GAKP01016822 GAKP01016821 JAC42131.1 KRG05647.1 KPU77623.1 GAMC01018839 JAB87716.1 KPU77621.1 KA649638 AFP64267.1 AGB93981.1 GEZM01048771 JAV76418.1 CH940682 KRF77567.1 JRES01000247 KNC33004.1 EAL29911.1 CH963876 EDW76855.1 GFDF01000153 JAV13931.1 GBXI01010096 JAD04196.1 OUUW01000009 SPP85249.1 EDW17619.1 GFDL01008932 JAV26113.1 CH480817 EDW50118.1 JXJN01008189 KRT08482.1 GGMR01013845 MBY26464.1 GFXV01007639 MBW19444.1 EDV38765.1 CH478619 EAT32709.1 KA646827 AFP61456.1 CP012525 ALC44812.1 GDHF01028761 GDHF01025122 GDHF01013738 GDHF01013593 GDHF01009620 GDHF01002806 JAI23553.1 JAI27192.1 JAI38576.1 JAI38721.1 JAI42694.1 JAI49508.1 GAKP01016823 GAKP01016820 JAC42129.1 AGB93982.1 EDW92906.1 EDV50227.1 JXUM01030164 JXUM01030165 GGMS01007914 MBY77117.1 EDW57089.1 GBXI01003963 JAD10329.1
Proteomes
UP000007151
UP000283053
UP000235965
UP000027135
UP000079169
UP000009046
+ More
UP000005205 UP000015103 UP000005203 UP000078492 UP000075809 UP000078540 UP000242457 UP000053097 UP000279307 UP000078542 UP000000311 UP000078541 UP000192223 UP000076502 UP000008237 UP000002358 UP000002282 UP000008711 UP000007266 UP000000803 UP000009192 UP000007801 UP000192221 UP000007755 UP000001819 UP000215335 UP000095301 UP000095300 UP000008792 UP000037069 UP000007798 UP000268350 UP000078200 UP000001292 UP000092460 UP000091820 UP000008820 UP000092553 UP000069940
UP000005205 UP000015103 UP000005203 UP000078492 UP000075809 UP000078540 UP000242457 UP000053097 UP000279307 UP000078542 UP000000311 UP000078541 UP000192223 UP000076502 UP000008237 UP000002358 UP000002282 UP000008711 UP000007266 UP000000803 UP000009192 UP000007801 UP000192221 UP000007755 UP000001819 UP000215335 UP000095301 UP000095300 UP000008792 UP000037069 UP000007798 UP000268350 UP000078200 UP000001292 UP000092460 UP000091820 UP000008820 UP000092553 UP000069940
Pfam
Interpro
ProteinModelPortal
A0A1E1WR33
A0A2H1VAD9
A0A173FDD0
S4PH37
A0A212FB96
A0A437BXV0
+ More
A0A2J7QVA1 A0A2J7QV98 A0A067RUI8 A0A069DXZ1 A0A1S3DD98 A0A224X4N5 A0A0V0G435 A0A2J7QVA8 A0A0K8SCT5 E0VV65 A0A0P4VTQ5 V9IJN9 A0A0K8TCJ0 A0A158NIL4 T1HPS9 A0A088ACE3 E9I8D6 A0A0C9R692 A0A151J4T0 A0A151XDV2 A0A195BCT0 A0A158NIL3 A0A2A3EFW3 A0A026W3N3 A0A151IP53 E2AX44 A0A195FLX6 A0A1W4XPA6 A0A411G645 A0A154P796 E2BFC6 K7J3F4 A0A1Y1LRY4 A0A0R1DUS3 A0A0Q5U4D6 D6W8J4 M9PDQ0 A0A0Q9XL73 A0A0P8ZSY5 A0A1W4W6C4 W8ANY7 F4WS96 A0A0R3P5Q6 A0A0Q5U2Q0 A0A0Q9XBE9 P13395 A0A232FF24 A0A0R1DZX8 A0A0Q5U8T3 A0A0R1DUG6 A0A1W4VTS2 A0A034VFQ9 A0A0Q9XAW4 A0A1I8ME62 A0A0P8Y852 W8B358 A0A0P9C1U7 T1PNU9 M9PGV6 A0A1Y1LU64 A0A1I8QDV7 A0A0Q9W9D3 A0A0L0CNI9 Q29EZ8 B4MXT6 A0A1L8E5B8 A0A0A1WZX6 A0A3B0KFZ2 B4KXU7 A0A1A9V2Y6 A0A1Q3FEY7 B4HW03 A0A1B0B477 A0A0R3P5T5 A0A2S2PAP5 A0A2H8TYT6 A0A1A9W748 A0A1I8ME59 B3M7A4 Q16EQ1 T1PDG2 A0A1I8ME61 A0A0M4F0U0 A0A0K8UAC6 A0A034VI07 M9PBI5 B4PDU1 B3NEV4 A0A1I8QDR3 A0A182H9F8 A0A1I8QDP4 A0A2S2QHA8 B4MGM3 A0A0A1XG25
A0A2J7QVA1 A0A2J7QV98 A0A067RUI8 A0A069DXZ1 A0A1S3DD98 A0A224X4N5 A0A0V0G435 A0A2J7QVA8 A0A0K8SCT5 E0VV65 A0A0P4VTQ5 V9IJN9 A0A0K8TCJ0 A0A158NIL4 T1HPS9 A0A088ACE3 E9I8D6 A0A0C9R692 A0A151J4T0 A0A151XDV2 A0A195BCT0 A0A158NIL3 A0A2A3EFW3 A0A026W3N3 A0A151IP53 E2AX44 A0A195FLX6 A0A1W4XPA6 A0A411G645 A0A154P796 E2BFC6 K7J3F4 A0A1Y1LRY4 A0A0R1DUS3 A0A0Q5U4D6 D6W8J4 M9PDQ0 A0A0Q9XL73 A0A0P8ZSY5 A0A1W4W6C4 W8ANY7 F4WS96 A0A0R3P5Q6 A0A0Q5U2Q0 A0A0Q9XBE9 P13395 A0A232FF24 A0A0R1DZX8 A0A0Q5U8T3 A0A0R1DUG6 A0A1W4VTS2 A0A034VFQ9 A0A0Q9XAW4 A0A1I8ME62 A0A0P8Y852 W8B358 A0A0P9C1U7 T1PNU9 M9PGV6 A0A1Y1LU64 A0A1I8QDV7 A0A0Q9W9D3 A0A0L0CNI9 Q29EZ8 B4MXT6 A0A1L8E5B8 A0A0A1WZX6 A0A3B0KFZ2 B4KXU7 A0A1A9V2Y6 A0A1Q3FEY7 B4HW03 A0A1B0B477 A0A0R3P5T5 A0A2S2PAP5 A0A2H8TYT6 A0A1A9W748 A0A1I8ME59 B3M7A4 Q16EQ1 T1PDG2 A0A1I8ME61 A0A0M4F0U0 A0A0K8UAC6 A0A034VI07 M9PBI5 B4PDU1 B3NEV4 A0A1I8QDR3 A0A182H9F8 A0A1I8QDP4 A0A2S2QHA8 B4MGM3 A0A0A1XG25
PDB
1U4Q
E-value=1.86585e-73,
Score=709
Ontologies
GO
GO:0005509
GO:0004725
GO:0005861
GO:0006937
GO:0051015
GO:0048306
GO:0003009
GO:0007417
GO:0005886
GO:0008360
GO:0045170
GO:0048790
GO:0007308
GO:0008017
GO:0007294
GO:0045478
GO:0007009
GO:0031594
GO:0030727
GO:0030707
GO:0048134
GO:0008092
GO:0016323
GO:0003779
GO:0042062
GO:0050807
GO:0005938
GO:0045169
GO:0005794
GO:0007274
GO:0051693
GO:0008091
GO:0007026
GO:0016199
GO:0005516
GO:0005515
GO:0006468
GO:0018024
GO:0007165
GO:0016620
GO:0006418
GO:0003676
GO:0016876
GO:0043039
Topology
Subcellular location
Cytoplasm
Cytoskeleton
Golgi apparatus
Cytoskeleton
Golgi apparatus
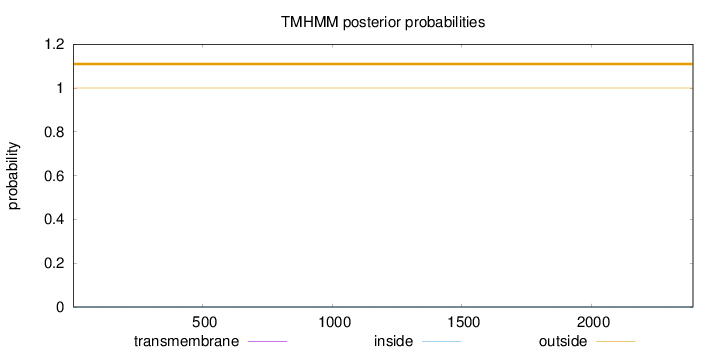
Length:
2392
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00023
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00002
outside
1 - 2392
Population Genetic Test Statistics
Pi
3.64528
Theta
19.126173
Tajima's D
-1.391803
CLR
169.198995
CSRT
0.0711464426778661
Interpretation
Uncertain
