Gene
KWMTBOMO08647
Pre Gene Modal
BGIBMGA007634
Annotation
PREDICTED:_dual_specificity_protein_kinase_TTK_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.038
Sequence
CDS
ATGAGTGGAAACACGGAATCTACTTCTAACAGCATCAAATTTGCTCCGATGCCAATGTCAGCTTTATTAGCTAATTTGATGCCTCTTCAAAAAGAGTCAGATACCCCTGATTCAAGTTTTTTGTCTGCTGACGAGAGCGATGATGATTCGATAGGCGAAACGGTCTCCAAGAAGCCTTCAACAAATAATAAAATAAATAGGGACTTCTACACACCTGTAATAAAACCCAAATCGAAAGAAGTTAAAGCGGTCGTTTTGGAGAAAAAAGCCTGTCCCACCTCTGAAGTGAAGAAACGTAATGTATTACTGCCATTTAATAACAGTACTGTTAAAAGTACTAATTCAGATATTAAACAAACACCTGAAATGATTAAAGTTCAAAATAGTGCACAGAAAAATAAATCTTCAACACCAGGGATTAAATCAGGTATACGTAAGTTTACACCAAACTCAGCTAAAAAGAGTCATAAGAAAACTCCACAGAACAGCATGATCCCGAGCAGAGATAGGGTGAGGTGTGAATTGTTTACGCAAAACAAAAAGGATGAGAAGAGCAATCAAATAGCACAGCCACAGCCAATAATACCTGAGCCGGAGACCCCCTTAAACAAAAAACTGATACCTGCATCATATGCAGCTACACCTTCATACCCTCAAGGCGCTGTAGGCAATAATTCCAAAATTTTATTTAAAACAACAAACATTAAGGATAAAAAGTACATGTTCATAAAGAAATTGGGCATAGGAGGTTCTAGTGAGGTTTACAAGGTCCTAGAAGTGGGAACATCTTGTGAGTATGCAGTAAAGTGTGTTTACTTAGCGACAGATCATGAACTGGCCCAAGGATATATCAATGAGGTTCGACTGCTCAGGGAATTGCAGAACTCAGACAGGGTTGTAAAGCTATATGACTATGAGTATGACAAATCACAACAGATGTTACGCATGGTTTTGGAAGTTGGTGAGACGGATTTATCAACATTCTTAAAAGGATGCAATTCGGGAGTACCTCCTGCTCTTGTGCTTCATTACTGGGAGGAAATGCTCCATGCTGTTAATTACATACATGAAAACGGTGTGATTCACGCAGACCTGAAGCCCGCAAACTTCTTGTTGGTGAGTGGACGTCTGAAGCTGATCGACTTCGGTATCGCGTCGTCGGTGAGCGGCGACGCGACGAGCGTGGTGCGGGCGCAAGCGGCCGGCACGTACTCCTACATCAGCCCCGAGGCGCTGCTCGGCGGCGCCGGACAACACGCGCACTCGCCGCCGGACGGCTCCGCGCCCATCAAGATCTCATTCCGTTCAGACGTGTGGTCTTTGGGCTGTATTCTGTACAGTATGGTTTATGGCCACACACCTTTTGCGCACATACGTAGTCTCGCTAAACTGGCTGCCATTCTCGACCCCAACCATCGGATAGAGTATCCGCCTGCTCCACATCTGCCTCCGTCGTTGCTGTCGACCCTGAAGTGGTGCCTGACGTACAACGCGCGAGGGCGGCCCTACGTGCGCGAGCTGCTGGCCGTGCAGCACCTGCAGCCGCCCAGCTCGCCGCTGCCGCAGCCGCTGCTCGACAAGCTGCAGAAGCTCGTCACGCCCCAGGAGTACAGCTTGCTGCAGAGAGCACAGATATGA
Protein
MSGNTESTSNSIKFAPMPMSALLANLMPLQKESDTPDSSFLSADESDDDSIGETVSKKPSTNNKINRDFYTPVIKPKSKEVKAVVLEKKACPTSEVKKRNVLLPFNNSTVKSTNSDIKQTPEMIKVQNSAQKNKSSTPGIKSGIRKFTPNSAKKSHKKTPQNSMIPSRDRVRCELFTQNKKDEKSNQIAQPQPIIPEPETPLNKKLIPASYAATPSYPQGAVGNNSKILFKTTNIKDKKYMFIKKLGIGGSSEVYKVLEVGTSCEYAVKCVYLATDHELAQGYINEVRLLRELQNSDRVVKLYDYEYDKSQQMLRMVLEVGETDLSTFLKGCNSGVPPALVLHYWEEMLHAVNYIHENGVIHADLKPANFLLVSGRLKLIDFGIASSVSGDATSVVRAQAAGTYSYISPEALLGGAGQHAHSPPDGSAPIKISFRSDVWSLGCILYSMVYGHTPFAHIRSLAKLAAILDPNHRIEYPPAPHLPPSLLSTLKWCLTYNARGRPYVRELLAVQHLQPPSSPLPQPLLDKLQKLVTPQEYSLLQRAQI
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
PDB
5NAD
E-value=2.11462e-50,
Score=504
Ontologies
GO
PANTHER
Topology
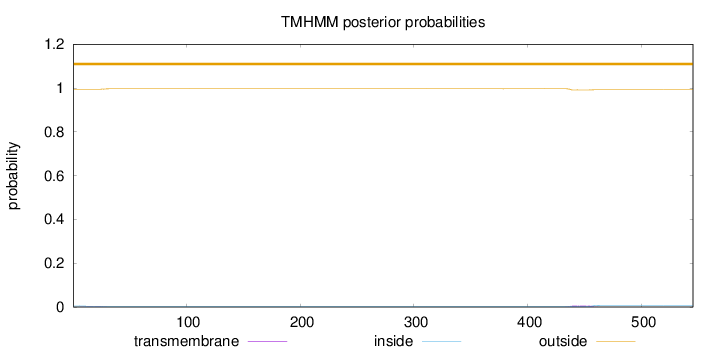
Length:
545
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15863
Exp number, first 60 AAs:
0.03072
Total prob of N-in:
0.00509
outside
1 - 545
Population Genetic Test Statistics
Pi
314.617632
Theta
224.164594
Tajima's D
1.30342
CLR
0
CSRT
0.740412979351032
Interpretation
Uncertain
