Gene
KWMTBOMO08643 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007949
Annotation
PREDICTED:_heat_shock_70_kD_protein_cognate_isoform_X1_[Bombyx_mori]
Full name
Delta-aminolevulinic acid dehydratase
Location in the cell
Cytoplasmic Reliability : 1.412 Mitochondrial Reliability : 1.406
Sequence
CDS
ATGAGTAATTTGTATTTAACACCTGAACATGTGCTGCAAGGCGGATACCATCATGCTGCCTTGAGAAAGTTACAGGAACCGAACACAAGCATCGAACCGCACAATATTATGTATCCTGTGTTTTTAGTAGAAAAAGATAATGCTATCGAACCGGTTCATAGTATGCCGAATGTAAACAGATATGGTGTTGACCAACTCATACCCCTTTTAGCAGAACTTGTGGAAAAAGGACTCAAATCTATACTCATTTTTGGGATTGTGGAGACACTGCCTAAGGACCCAACAGGTACTAGTGCAGATTCAGTAGACAATCCAGTAATAAAAGCATTGCCGAAATTGCGTGCTGCCTTTCCTGATTTGTTAATAGCATGTGATGTATGTTTATGTCCGTACACATCACACGGACATTGTGGCTTACTGAAGAGTGGAGGAGGTATCGACCATGAAGCATCAGTAAAGAGGATAGCTGAAGTCGCATTGGCTTATGCAAAAGCTGGTGCTCATGTGGTGGCTCCTTCAGACATGATGGACAATAGGATTAAAGCAATCAAAGAGGAGCTGGTCAGAAATAAATTGCAAAACCAAGTGTCGATACTGTCTTACTCGTGTAAGTTCGCGTCGTCGATGTACGGACCCTTCCGTGACACTATGAAGAGCTCTCCGATGGAAGGCGACCGGAAATGCTATCAGCTTCCGCCGGGCAGCGCCGCTTTAGCCGCCAGGGCCGCAGCGCGTGACGTTGCTGAAGGCGCAGACTTCCTGATGGTGAAACCGGGGCTGCCCTACCTGGACATTGTGCGGCAGACCAAGGACCGCTTCCCGCACCACCCCTTGTTCATATACCAGGTATCCGGCGAGTACGAGTTGATATCTAGGAGCGGTGACCGCGCGAGCATGCAGACCAACCTGATGGAGATCCTCACGTGCATGAGGAGGGCGGGCGCTGATTGCATCATAACGTATTTCGCTCCTCTCATTTTAAGCATAATATCAGAAAAGAAGTAA
Protein
MSNLYLTPEHVLQGGYHHAALRKLQEPNTSIEPHNIMYPVFLVEKDNAIEPVHSMPNVNRYGVDQLIPLLAELVEKGLKSILIFGIVETLPKDPTGTSADSVDNPVIKALPKLRAAFPDLLIACDVCLCPYTSHGHCGLLKSGGGIDHEASVKRIAEVALAYAKAGAHVVAPSDMMDNRIKAIKEELVRNKLQNQVSILSYSCKFASSMYGPFRDTMKSSPMEGDRKCYQLPPGSAALAARAAARDVAEGADFLMVKPGLPYLDIVRQTKDRFPHHPLFIYQVSGEYELISRSGDRASMQTNLMEILTCMRRAGADCIITYFAPLILSIISEKK
Summary
Catalytic Activity
2 5-aminolevulinate = H(+) + 2 H2O + porphobilinogen
Subunit
Homooctamer.
Similarity
Belongs to the ALAD family.
Uniprot
S4PGN8
A0A2A4K674
A0A3S2TU09
A0A212FBA0
A0A2H1WWZ8
H9JEK2
+ More
A0A0K8TSS3 A0A182V005 B0WF62 Q1HQH0 A0A1Q3FC04 A0A182KKC0 A0A182LZ53 A0A1B0CG52 A0A182WUN9 Q7QGX8 A0A182IZU1 A0A182RYW3 A0A182HVT6 A0A182QJH4 A0A182PJA3 A0A182UIS9 A0A182GZ32 A0A182K5A2 A0A2M4BSG8 A0A182FKD1 A0A182GYT3 A0A2M3ZGX4 A0A2M4ASP0 W5JNK1 A0A2M4CG71 A0A182WFW4 A0A182YKM8 T1E9Y9 A0A182SJR4 A0A195DMQ8 A0A0N7ZCW9 K7ITP7 A0A0L0BWK3 T1PIQ7 A0A151IKJ1 U5EKG4 A0A195FJL2 F4W674 R4G3N4 A0A0C9RCB1 A0A034WPC6 A0A310SGR9 A0A423SST2 A0A1W4WIU1 A0A195AW61 A0A0K8VLB7 A0A1I8P405 W8CCQ8 A0A336LML7 J3JVW8 A0A0A9XHP9 A0A0K8VZ16 A0A0A9XN56 A0A154PI30 A0A0A9XJW8 A0A0B7ANN1 A0A026VU46 A0A0L7R5D2 A0A1A9WCZ5 A0A0A1XEV8 A0A0J7L3H2 V5GG21 A0A067RKI6 E0VHL1 A0A194PW64 A0A069DYN0 B4HFK9 A0A088ACF3 B3NH88 B4QR82 A0A084VSI6 B4PGB6 A0A2A3EH53 Q2M0R1 A0A224XJP0 B4GR82 B4MLG5 Q9VTV9 R7TCP0 B3M7U1 D0QWS1 A0A1B0C5L0 A0A1Y1JY47 A0A0V0G5V8 B4LFF2 A0A1S3K0K7 A0A1A9XPZ7 N6TWI0 A0A444TBC1 B4KZZ5 A0A3B0J4A6 A0A210Q6E1 H2TPX9 A0A0M5J151
A0A0K8TSS3 A0A182V005 B0WF62 Q1HQH0 A0A1Q3FC04 A0A182KKC0 A0A182LZ53 A0A1B0CG52 A0A182WUN9 Q7QGX8 A0A182IZU1 A0A182RYW3 A0A182HVT6 A0A182QJH4 A0A182PJA3 A0A182UIS9 A0A182GZ32 A0A182K5A2 A0A2M4BSG8 A0A182FKD1 A0A182GYT3 A0A2M3ZGX4 A0A2M4ASP0 W5JNK1 A0A2M4CG71 A0A182WFW4 A0A182YKM8 T1E9Y9 A0A182SJR4 A0A195DMQ8 A0A0N7ZCW9 K7ITP7 A0A0L0BWK3 T1PIQ7 A0A151IKJ1 U5EKG4 A0A195FJL2 F4W674 R4G3N4 A0A0C9RCB1 A0A034WPC6 A0A310SGR9 A0A423SST2 A0A1W4WIU1 A0A195AW61 A0A0K8VLB7 A0A1I8P405 W8CCQ8 A0A336LML7 J3JVW8 A0A0A9XHP9 A0A0K8VZ16 A0A0A9XN56 A0A154PI30 A0A0A9XJW8 A0A0B7ANN1 A0A026VU46 A0A0L7R5D2 A0A1A9WCZ5 A0A0A1XEV8 A0A0J7L3H2 V5GG21 A0A067RKI6 E0VHL1 A0A194PW64 A0A069DYN0 B4HFK9 A0A088ACF3 B3NH88 B4QR82 A0A084VSI6 B4PGB6 A0A2A3EH53 Q2M0R1 A0A224XJP0 B4GR82 B4MLG5 Q9VTV9 R7TCP0 B3M7U1 D0QWS1 A0A1B0C5L0 A0A1Y1JY47 A0A0V0G5V8 B4LFF2 A0A1S3K0K7 A0A1A9XPZ7 N6TWI0 A0A444TBC1 B4KZZ5 A0A3B0J4A6 A0A210Q6E1 H2TPX9 A0A0M5J151
EC Number
4.2.1.24
Pubmed
23622113
22118469
19121390
26369729
17204158
17510324
+ More
20966253 12364791 14747013 17210077 26483478 20920257 23761445 25244985 20075255 26108605 25315136 21719571 25348373 24495485 22516182 25401762 26823975 24508170 30249741 25830018 24845553 20566863 26354079 26334808 17994087 22936249 24438588 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23254933 19859648 28004739 23537049 28812685 21551351
20966253 12364791 14747013 17210077 26483478 20920257 23761445 25244985 20075255 26108605 25315136 21719571 25348373 24495485 22516182 25401762 26823975 24508170 30249741 25830018 24845553 20566863 26354079 26334808 17994087 22936249 24438588 17550304 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23254933 19859648 28004739 23537049 28812685 21551351
EMBL
GAIX01003577
JAA88983.1
NWSH01000087
PCG79737.1
RSAL01000001
RVE55213.1
+ More
AGBW02009363 OWR50997.1 ODYU01011696 SOQ57599.1 BABH01003235 BABH01003236 BABH01003237 GDAI01000191 JAI17412.1 DS231915 EDS26038.1 DQ440474 CH477238 ABF18507.1 EAT46522.1 GFDL01010003 JAV25042.1 AXCM01007054 AJWK01010758 AAAB01008823 EAA05538.4 AXCP01008426 APCN01001416 AXCN02001762 JXUM01021456 KQ560601 KXJ81683.1 GGFJ01006793 MBW55934.1 JXUM01098060 KQ564391 KXJ72354.1 GGFM01006974 MBW27725.1 GGFK01010495 MBW43816.1 ADMH02001012 ETN64460.1 GGFL01000136 MBW64314.1 GAMD01000877 JAB00714.1 KQ980724 KYN14116.1 GDRN01058260 JAI65616.1 JRES01001238 KNC24408.1 KA647985 AFP62614.1 KQ977199 KYN05001.1 GANO01001822 JAB58049.1 KQ981523 KYN40447.1 GL887707 EGI70209.1 GAHY01001511 JAA75999.1 GBYB01014214 JAG83981.1 GAKP01001516 GAKP01001515 JAC57436.1 KQ760132 OAD61810.1 QCYY01002829 ROT67257.1 KQ976731 KYM76292.1 GDHF01012645 JAI39669.1 GAMC01003271 JAC03285.1 UFQT01000011 SSX17597.1 BT127386 AEE62348.1 GBHO01023347 JAG20257.1 GDHF01008156 JAI44158.1 GBHO01023349 GBHO01023346 GBRD01007230 JAG20255.1 JAG20258.1 JAG58591.1 KQ434923 KZC11499.1 GBHO01023345 GDHC01012839 JAG20259.1 JAQ05790.1 HACG01034640 CEK81505.1 KK111518 QOIP01000005 EZA46379.1 RLU23142.1 KQ414654 KOC66029.1 GBXI01004795 JAD09497.1 LBMM01000815 KMQ97427.1 GALX01005552 JAB62914.1 KK852460 KDR23528.1 DS235171 EEB12897.1 KQ459596 KPI95375.1 GBGD01002105 JAC86784.1 CH480815 EDW41240.1 CH954178 EDV51545.1 CM000363 CM002912 EDX10212.1 KMY99204.1 ATLV01016038 KE525054 KFB40930.1 CM000159 EDW94280.1 KZ288256 PBC30612.1 CH379069 EAL30869.2 GFTR01005162 JAW11264.1 CH479188 EDW40267.1 CH963847 EDW72821.1 AE014296 AY071047 AAF49936.1 AAL48669.1 AGB94445.1 AMQN01002930 KB310466 ELT91513.1 CH902618 EDV38814.1 FJ821035 ACN94783.1 JXJN01026061 GEZM01097362 JAV54252.1 GECL01002739 JAP03385.1 CH940647 EDW69250.1 APGK01049750 KB741156 KB631941 ENN73615.1 ERL87379.1 SAUD01005232 RXG67273.1 CH933809 EDW18007.1 OUUW01000002 SPP76285.1 NEDP02004840 OWF44259.1 CP012525 ALC45061.1
AGBW02009363 OWR50997.1 ODYU01011696 SOQ57599.1 BABH01003235 BABH01003236 BABH01003237 GDAI01000191 JAI17412.1 DS231915 EDS26038.1 DQ440474 CH477238 ABF18507.1 EAT46522.1 GFDL01010003 JAV25042.1 AXCM01007054 AJWK01010758 AAAB01008823 EAA05538.4 AXCP01008426 APCN01001416 AXCN02001762 JXUM01021456 KQ560601 KXJ81683.1 GGFJ01006793 MBW55934.1 JXUM01098060 KQ564391 KXJ72354.1 GGFM01006974 MBW27725.1 GGFK01010495 MBW43816.1 ADMH02001012 ETN64460.1 GGFL01000136 MBW64314.1 GAMD01000877 JAB00714.1 KQ980724 KYN14116.1 GDRN01058260 JAI65616.1 JRES01001238 KNC24408.1 KA647985 AFP62614.1 KQ977199 KYN05001.1 GANO01001822 JAB58049.1 KQ981523 KYN40447.1 GL887707 EGI70209.1 GAHY01001511 JAA75999.1 GBYB01014214 JAG83981.1 GAKP01001516 GAKP01001515 JAC57436.1 KQ760132 OAD61810.1 QCYY01002829 ROT67257.1 KQ976731 KYM76292.1 GDHF01012645 JAI39669.1 GAMC01003271 JAC03285.1 UFQT01000011 SSX17597.1 BT127386 AEE62348.1 GBHO01023347 JAG20257.1 GDHF01008156 JAI44158.1 GBHO01023349 GBHO01023346 GBRD01007230 JAG20255.1 JAG20258.1 JAG58591.1 KQ434923 KZC11499.1 GBHO01023345 GDHC01012839 JAG20259.1 JAQ05790.1 HACG01034640 CEK81505.1 KK111518 QOIP01000005 EZA46379.1 RLU23142.1 KQ414654 KOC66029.1 GBXI01004795 JAD09497.1 LBMM01000815 KMQ97427.1 GALX01005552 JAB62914.1 KK852460 KDR23528.1 DS235171 EEB12897.1 KQ459596 KPI95375.1 GBGD01002105 JAC86784.1 CH480815 EDW41240.1 CH954178 EDV51545.1 CM000363 CM002912 EDX10212.1 KMY99204.1 ATLV01016038 KE525054 KFB40930.1 CM000159 EDW94280.1 KZ288256 PBC30612.1 CH379069 EAL30869.2 GFTR01005162 JAW11264.1 CH479188 EDW40267.1 CH963847 EDW72821.1 AE014296 AY071047 AAF49936.1 AAL48669.1 AGB94445.1 AMQN01002930 KB310466 ELT91513.1 CH902618 EDV38814.1 FJ821035 ACN94783.1 JXJN01026061 GEZM01097362 JAV54252.1 GECL01002739 JAP03385.1 CH940647 EDW69250.1 APGK01049750 KB741156 KB631941 ENN73615.1 ERL87379.1 SAUD01005232 RXG67273.1 CH933809 EDW18007.1 OUUW01000002 SPP76285.1 NEDP02004840 OWF44259.1 CP012525 ALC45061.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000005204
UP000075903
UP000002320
+ More
UP000008820 UP000075882 UP000075883 UP000092461 UP000076407 UP000007062 UP000075880 UP000075900 UP000075840 UP000075886 UP000075885 UP000075902 UP000069940 UP000249989 UP000075881 UP000069272 UP000000673 UP000075920 UP000076408 UP000075901 UP000078492 UP000002358 UP000037069 UP000095301 UP000078542 UP000078541 UP000007755 UP000283509 UP000192223 UP000078540 UP000095300 UP000076502 UP000053097 UP000279307 UP000053825 UP000091820 UP000036403 UP000027135 UP000009046 UP000053268 UP000001292 UP000005203 UP000008711 UP000000304 UP000030765 UP000002282 UP000242457 UP000001819 UP000008744 UP000007798 UP000000803 UP000014760 UP000007801 UP000092460 UP000008792 UP000085678 UP000092443 UP000019118 UP000030742 UP000288706 UP000009192 UP000268350 UP000242188 UP000005226 UP000092553
UP000008820 UP000075882 UP000075883 UP000092461 UP000076407 UP000007062 UP000075880 UP000075900 UP000075840 UP000075886 UP000075885 UP000075902 UP000069940 UP000249989 UP000075881 UP000069272 UP000000673 UP000075920 UP000076408 UP000075901 UP000078492 UP000002358 UP000037069 UP000095301 UP000078542 UP000078541 UP000007755 UP000283509 UP000192223 UP000078540 UP000095300 UP000076502 UP000053097 UP000279307 UP000053825 UP000091820 UP000036403 UP000027135 UP000009046 UP000053268 UP000001292 UP000005203 UP000008711 UP000000304 UP000030765 UP000002282 UP000242457 UP000001819 UP000008744 UP000007798 UP000000803 UP000014760 UP000007801 UP000092460 UP000008792 UP000085678 UP000092443 UP000019118 UP000030742 UP000288706 UP000009192 UP000268350 UP000242188 UP000005226 UP000092553
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
S4PGN8
A0A2A4K674
A0A3S2TU09
A0A212FBA0
A0A2H1WWZ8
H9JEK2
+ More
A0A0K8TSS3 A0A182V005 B0WF62 Q1HQH0 A0A1Q3FC04 A0A182KKC0 A0A182LZ53 A0A1B0CG52 A0A182WUN9 Q7QGX8 A0A182IZU1 A0A182RYW3 A0A182HVT6 A0A182QJH4 A0A182PJA3 A0A182UIS9 A0A182GZ32 A0A182K5A2 A0A2M4BSG8 A0A182FKD1 A0A182GYT3 A0A2M3ZGX4 A0A2M4ASP0 W5JNK1 A0A2M4CG71 A0A182WFW4 A0A182YKM8 T1E9Y9 A0A182SJR4 A0A195DMQ8 A0A0N7ZCW9 K7ITP7 A0A0L0BWK3 T1PIQ7 A0A151IKJ1 U5EKG4 A0A195FJL2 F4W674 R4G3N4 A0A0C9RCB1 A0A034WPC6 A0A310SGR9 A0A423SST2 A0A1W4WIU1 A0A195AW61 A0A0K8VLB7 A0A1I8P405 W8CCQ8 A0A336LML7 J3JVW8 A0A0A9XHP9 A0A0K8VZ16 A0A0A9XN56 A0A154PI30 A0A0A9XJW8 A0A0B7ANN1 A0A026VU46 A0A0L7R5D2 A0A1A9WCZ5 A0A0A1XEV8 A0A0J7L3H2 V5GG21 A0A067RKI6 E0VHL1 A0A194PW64 A0A069DYN0 B4HFK9 A0A088ACF3 B3NH88 B4QR82 A0A084VSI6 B4PGB6 A0A2A3EH53 Q2M0R1 A0A224XJP0 B4GR82 B4MLG5 Q9VTV9 R7TCP0 B3M7U1 D0QWS1 A0A1B0C5L0 A0A1Y1JY47 A0A0V0G5V8 B4LFF2 A0A1S3K0K7 A0A1A9XPZ7 N6TWI0 A0A444TBC1 B4KZZ5 A0A3B0J4A6 A0A210Q6E1 H2TPX9 A0A0M5J151
A0A0K8TSS3 A0A182V005 B0WF62 Q1HQH0 A0A1Q3FC04 A0A182KKC0 A0A182LZ53 A0A1B0CG52 A0A182WUN9 Q7QGX8 A0A182IZU1 A0A182RYW3 A0A182HVT6 A0A182QJH4 A0A182PJA3 A0A182UIS9 A0A182GZ32 A0A182K5A2 A0A2M4BSG8 A0A182FKD1 A0A182GYT3 A0A2M3ZGX4 A0A2M4ASP0 W5JNK1 A0A2M4CG71 A0A182WFW4 A0A182YKM8 T1E9Y9 A0A182SJR4 A0A195DMQ8 A0A0N7ZCW9 K7ITP7 A0A0L0BWK3 T1PIQ7 A0A151IKJ1 U5EKG4 A0A195FJL2 F4W674 R4G3N4 A0A0C9RCB1 A0A034WPC6 A0A310SGR9 A0A423SST2 A0A1W4WIU1 A0A195AW61 A0A0K8VLB7 A0A1I8P405 W8CCQ8 A0A336LML7 J3JVW8 A0A0A9XHP9 A0A0K8VZ16 A0A0A9XN56 A0A154PI30 A0A0A9XJW8 A0A0B7ANN1 A0A026VU46 A0A0L7R5D2 A0A1A9WCZ5 A0A0A1XEV8 A0A0J7L3H2 V5GG21 A0A067RKI6 E0VHL1 A0A194PW64 A0A069DYN0 B4HFK9 A0A088ACF3 B3NH88 B4QR82 A0A084VSI6 B4PGB6 A0A2A3EH53 Q2M0R1 A0A224XJP0 B4GR82 B4MLG5 Q9VTV9 R7TCP0 B3M7U1 D0QWS1 A0A1B0C5L0 A0A1Y1JY47 A0A0V0G5V8 B4LFF2 A0A1S3K0K7 A0A1A9XPZ7 N6TWI0 A0A444TBC1 B4KZZ5 A0A3B0J4A6 A0A210Q6E1 H2TPX9 A0A0M5J151
PDB
2Z1B
E-value=2.79901e-97,
Score=906
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Chromosome
Nucleus
Nucleus
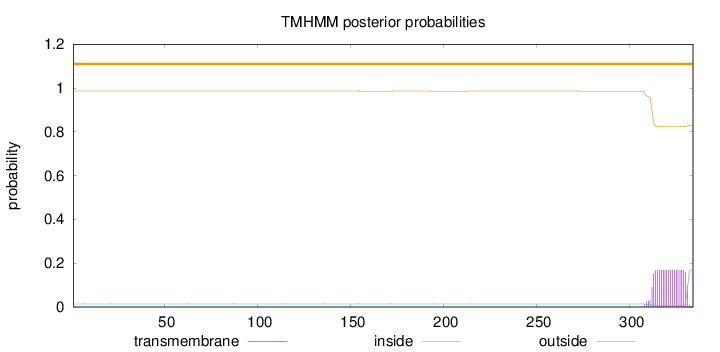
Length:
334
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.31926
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01405
outside
1 - 334
Population Genetic Test Statistics
Pi
328.578181
Theta
179.0156
Tajima's D
-1.980895
CLR
1552.611032
CSRT
0.0144992750362482
Interpretation
Uncertain
