Gene
KWMTBOMO08641
Pre Gene Modal
BGIBMGA007633
Annotation
PREDICTED:_adenylate_kinase_8-like_isoform_X2_[Bombyx_mori]
Full name
Adenylate kinase 8
Alternative Name
ATP-AMP transphosphorylase 8
Location in the cell
Cytoplasmic Reliability : 1.327 Nuclear Reliability : 1.773
Sequence
CDS
ATGGAAAAATATAGAGTATACCAAATATTTAAGGACATGTTGAAAGATCTACTAATCAACTTGCCCAAAGATCATTTGAAGCACATGAAAATATTCTTACATCGTTATTCGCAGAGCTGTAAAGATGCGGATAGGATCATCCTATTAGTATCACCAGAGCTCGAACATATCGATATCACAGCCATTCTAAAACAGTTGATAAAGGACTTGGGTTTGTTCGTGATCACACGCCGAGTCGTAATGGATCGATACGAGAAGCACGAAAATTACATGCCCGGATGCGTGTCTCCTGTTGTCTTATCGGAAATTACGAAAACTCTTACTATGAAAGATCAAATACTTCAAGCCGGCTGGTTGATGTTTGACCATCCCTGCACAGTGCGGGAGGCTCGTTGTCTGCAGCAGGACGGTGTGCTGCCAACCGTGACACTAGCCCTCATGCCAACCCCCCCTCAAGCTCCTCCGTCTGACAATCTACTCACATCAGCTAGATCGTTTTTTCAGCAGGATTTCGAAGGACTCAAGTTTGCATACAAGGCTACTCTAAAGGAGGTGTACATTGATTCCGAAGATGAAATAGAAAAAATATCAACAAAATGCTTTAATGCCATTCGGGCGTGTGCGGCCGGCGCTCAAGGACCATACCAGGGACACCACGTGGTCGGCGCTCCTGGAGTGTACAGAGTACTTGTTATTGGACCTCGTGGATCGGGCCGCAGGTCTCAGGCTTTGGCTCTGGCCAAACATTTCCGCTTAGTCTATTTACATTTCGACGATTTGTTAAGTGAAGCTAAAGCAAAGACCGACGAGTTTGGACAGAAACTACGTCAACATGAACCGAGCGTGCAAGTGAAAGCTGTCATAGTGAAGCGACGTATCTTGTTGAAGGACTGCATCGATAACGGTTGGGTTCTCGTTGGATTCCCGATGTCGGGAAAAGAATTCGAACTTTTAGACAATACACCGACCCCGCCAAATAGAATAATAATATTGAACGTGGATGCTTCAACGTGTAAGAGTCGTCGCTTGAGTCGAGGGGTGGACTGGTGCAGCGGTGATGAAGCACCCTTGGGTTCAGGGTCTCGTGTTATGCGGCATCCTAGAGATGACGAAGCTCAACTTGAAACCGAGTTAGACAACTATTTCTCCGAGGCATTAGCGGAGCTACGAGCGGCGGCCGGCATCACTGCGGTGGAATTCGACGCTAGTCAATCTTTTGACGGCGTGCAAGTGAAACTGCAGGCGGCCGTGATGGCGGCACCTAGTTTTGATATAGAGTGCGGGGCGCAACTCTGCCACGTCCGAGCAGACTGA
Protein
MEKYRVYQIFKDMLKDLLINLPKDHLKHMKIFLHRYSQSCKDADRIILLVSPELEHIDITAILKQLIKDLGLFVITRRVVMDRYEKHENYMPGCVSPVVLSEITKTLTMKDQILQAGWLMFDHPCTVREARCLQQDGVLPTVTLALMPTPPQAPPSDNLLTSARSFFQQDFEGLKFAYKATLKEVYIDSEDEIEKISTKCFNAIRACAAGAQGPYQGHHVVGAPGVYRVLVIGPRGSGRRSQALALAKHFRLVYLHFDDLLSEAKAKTDEFGQKLRQHEPSVQVKAVIVKRRILLKDCIDNGWVLVGFPMSGKEFELLDNTPTPPNRIIILNVDASTCKSRRLSRGVDWCSGDEAPLGSGSRVMRHPRDDEAQLETELDNYFSEALAELRAAAGITAVEFDASQSFDGVQVKLQAAVMAAPSFDIECGAQLCHVRAD
Summary
Description
Nucleoside monophosphate (NMP) kinase that catalyzes the reversible transfer of the terminal phosphate group between nucleoside triphosphates and monophosphates. Has highest activity toward AMP, and weaker activity toward dAMP, CMP and dCMP. Also displays broad nucleoside diphosphate kinase activity.
Catalytic Activity
AMP + ATP = 2 ADP
a 2'-deoxyribonucleoside 5'-diphosphate + ATP = a 2'-deoxyribonucleoside 5'-triphosphate + ADP
a ribonucleoside 5'-diphosphate + ATP = a ribonucleoside 5'-triphosphate + ADP
a 2'-deoxyribonucleoside 5'-diphosphate + ATP = a 2'-deoxyribonucleoside 5'-triphosphate + ADP
a ribonucleoside 5'-diphosphate + ATP = a ribonucleoside 5'-triphosphate + ADP
Biophysicochemical Properties
4.1 uM for AMP
630 uM for dAMP
1.4 uM for CMP
630 uM for dAMP
1.4 uM for CMP
Similarity
Belongs to the adenylate kinase family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Kinase
Nucleotide-binding
Polymorphism
Reference proteome
Transferase
Feature
chain Adenylate kinase 8
splice variant In isoform 2.
sequence variant In dbSNP:rs2231400.
splice variant In isoform 2.
sequence variant In dbSNP:rs2231400.
Uniprot
H9JDN6
A0A194PPY0
A0A437BXM4
A0A0N0PD18
A0A2A4K7V9
A0A0L7L2F2
+ More
A0A139WMM8 E9J5Q4 A0A0L7R566 A0A2A3EFP1 A0A088AK29 A0A195CI11 A0A310SVP4 A0A195F0M6 A0A195E742 A0A151XBE6 A0A154PI47 A0A1B6CVM8 U4UKA9 N6TH76 A0A151I186 A0A1B6L896 A0A026VVQ7 J9LPR0 A0A1B6FAH0 A0A0A9Y5J0 A0A224XAJ8 A0A232EIA6 A0A2S2NR53 A0A452SIU4 A0A3Q7Y1A4 A0A2U3VDI1 H2PTS5 A0A3Q7TVT5 G1LBZ8 A0A3Q7RUD3 U6CW53 E2QSP0 A0A2Y9EH63 H0Y2C7 A0A2Y9H6C9 A0A3P4QBT3 A0A2J8UJ55 A0A2K6NBZ3 A0A2K6KU67 H2QY34 A0A2R9B4R4 A0A2J8NAJ2 Q96MA6 M3YM49 A0A2K6FVR1 A0A2K6TA67 U3CCA3 A0A2K6E6S5 A0A2K5JYB2 H9EWJ8 A0A096P0D8 A0A2K5M076 G7PRA1 F6YCP3 A0A0D9RR30 F6PLL4 A0A2Y9J8U8 Q4R3W4 A0A1S3A1N3 A0A2I2V3K2 Q502L7 L5K726 U3I3A1 A0A2Y9DJY9 A2BFD3 A0A1A8FDG0 U3JFI3 I3M2A2 A0A2R8Q451 A0A3M7QQ56 A0A2K5D2I4 A0A3B3RYF1 A0A0B8RRT5 Q68FP8 E1BLG0 A0A452UP54 A0A401PII7 Q32M07 A0A1Y1XJS7 A0A1D5PVQ4
A0A139WMM8 E9J5Q4 A0A0L7R566 A0A2A3EFP1 A0A088AK29 A0A195CI11 A0A310SVP4 A0A195F0M6 A0A195E742 A0A151XBE6 A0A154PI47 A0A1B6CVM8 U4UKA9 N6TH76 A0A151I186 A0A1B6L896 A0A026VVQ7 J9LPR0 A0A1B6FAH0 A0A0A9Y5J0 A0A224XAJ8 A0A232EIA6 A0A2S2NR53 A0A452SIU4 A0A3Q7Y1A4 A0A2U3VDI1 H2PTS5 A0A3Q7TVT5 G1LBZ8 A0A3Q7RUD3 U6CW53 E2QSP0 A0A2Y9EH63 H0Y2C7 A0A2Y9H6C9 A0A3P4QBT3 A0A2J8UJ55 A0A2K6NBZ3 A0A2K6KU67 H2QY34 A0A2R9B4R4 A0A2J8NAJ2 Q96MA6 M3YM49 A0A2K6FVR1 A0A2K6TA67 U3CCA3 A0A2K6E6S5 A0A2K5JYB2 H9EWJ8 A0A096P0D8 A0A2K5M076 G7PRA1 F6YCP3 A0A0D9RR30 F6PLL4 A0A2Y9J8U8 Q4R3W4 A0A1S3A1N3 A0A2I2V3K2 Q502L7 L5K726 U3I3A1 A0A2Y9DJY9 A2BFD3 A0A1A8FDG0 U3JFI3 I3M2A2 A0A2R8Q451 A0A3M7QQ56 A0A2K5D2I4 A0A3B3RYF1 A0A0B8RRT5 Q68FP8 E1BLG0 A0A452UP54 A0A401PII7 Q32M07 A0A1Y1XJS7 A0A1D5PVQ4
EC Number
2.7.4.3
2.7.4.6
2.7.4.6
Pubmed
19121390
26354079
26227816
18362917
19820115
21282665
+ More
23537049 24508170 25401762 28648823 20010809 16341006 25362486 16136131 22722832 14702039 15164053 15489334 21080915 23416111 25243066 25319552 22002653 17431167 17495919 17975172 23258410 23749191 23594743 30375419 29240929 25476704 19393038 24813606 30297745 19468303 21183079 15592404
23537049 24508170 25401762 28648823 20010809 16341006 25362486 16136131 22722832 14702039 15164053 15489334 21080915 23416111 25243066 25319552 22002653 17431167 17495919 17975172 23258410 23749191 23594743 30375419 29240929 25476704 19393038 24813606 30297745 19468303 21183079 15592404
EMBL
BABH01003237
KQ459596
KPI95372.1
RSAL01000001
RVE55215.1
KQ460497
+ More
KPJ14330.1 NWSH01000087 PCG79740.1 JTDY01003375 KOB69653.1 KQ971312 KYB29097.1 GL768174 EFZ11860.1 KQ414654 KOC65988.1 KZ288256 PBC30575.1 KQ977721 KYN00371.1 KQ759867 OAD62463.1 KQ981864 KYN34135.1 KQ979568 KYN21008.1 KQ982335 KYQ57664.1 KQ434923 KZC11525.1 GEDC01020025 JAS17273.1 KB632338 ERL92898.1 APGK01038386 KB740954 ENN77093.1 KQ976580 KYM79962.1 GEBQ01020057 JAT19920.1 KK107766 EZA47873.1 ABLF02028091 GECZ01022624 JAS47145.1 GBHO01015277 GBRD01002305 JAG28327.1 JAG63516.1 GFTR01007020 JAW09406.1 NNAY01004284 OXU18107.1 GGMR01006988 MBY19607.1 ABGA01088221 ABGA01088222 ABGA01088223 ABGA01088224 ABGA01088225 ABGA01088226 ABGA01088227 ABGA01088228 ABGA01088229 ABGA01088230 ACTA01018302 ACTA01026302 ACTA01034302 ACTA01042301 HAAF01002713 CCP74539.1 AAEX03006812 AAEX03006813 AAQR03116685 AAQR03116686 AAQR03116687 AAQR03116688 AAQR03116689 AAQR03116690 CYRY02036932 VCX20087.1 NDHI03003456 PNJ45289.1 AACZ04069149 AJFE02078021 AJFE02078022 AJFE02078023 AJFE02078024 AJFE02078025 AJFE02078026 AJFE02078027 AJFE02078028 AJFE02078029 NBAG03000232 PNI68780.1 AK057266 AK093446 AK292186 AL160165 AL445645 CH471090 BC034776 BC050576 AEYP01042405 AEYP01042406 AEYP01042407 AEYP01042408 AEYP01042409 AEYP01042410 AEYP01042411 GAMS01006604 GAMP01003350 JAB16532.1 JAB49405.1 JU323001 AFE66757.1 AHZZ02007974 AQIA01023155 AQIA01023156 AQIA01023157 AQIA01023158 AQIA01023159 CM001290 EHH57017.1 JSUE03013934 CM001267 EHH23721.1 AQIB01140538 AQIB01140539 AQIB01140540 AQIB01140541 AQIB01140542 AQIB01140543 AQIB01140544 AQIB01140545 AB179151 AANG04002451 BC095649 KB030979 ELK07315.1 ADON01031071 ADON01031072 ADON01031073 BX276128 HAEB01010306 SBQ56833.1 AGTO01010822 AGTP01020290 AGTP01020291 AGTP01020292 BX510918 REGN01005372 RNA13587.1 GBSH01000210 JAG68814.1 BC079446 BFAA01000555 GCB72946.1 AL731851 AL732526 BC100481 BC109352 MCFG01000027 ORX85975.1 AADN05000150
KPJ14330.1 NWSH01000087 PCG79740.1 JTDY01003375 KOB69653.1 KQ971312 KYB29097.1 GL768174 EFZ11860.1 KQ414654 KOC65988.1 KZ288256 PBC30575.1 KQ977721 KYN00371.1 KQ759867 OAD62463.1 KQ981864 KYN34135.1 KQ979568 KYN21008.1 KQ982335 KYQ57664.1 KQ434923 KZC11525.1 GEDC01020025 JAS17273.1 KB632338 ERL92898.1 APGK01038386 KB740954 ENN77093.1 KQ976580 KYM79962.1 GEBQ01020057 JAT19920.1 KK107766 EZA47873.1 ABLF02028091 GECZ01022624 JAS47145.1 GBHO01015277 GBRD01002305 JAG28327.1 JAG63516.1 GFTR01007020 JAW09406.1 NNAY01004284 OXU18107.1 GGMR01006988 MBY19607.1 ABGA01088221 ABGA01088222 ABGA01088223 ABGA01088224 ABGA01088225 ABGA01088226 ABGA01088227 ABGA01088228 ABGA01088229 ABGA01088230 ACTA01018302 ACTA01026302 ACTA01034302 ACTA01042301 HAAF01002713 CCP74539.1 AAEX03006812 AAEX03006813 AAQR03116685 AAQR03116686 AAQR03116687 AAQR03116688 AAQR03116689 AAQR03116690 CYRY02036932 VCX20087.1 NDHI03003456 PNJ45289.1 AACZ04069149 AJFE02078021 AJFE02078022 AJFE02078023 AJFE02078024 AJFE02078025 AJFE02078026 AJFE02078027 AJFE02078028 AJFE02078029 NBAG03000232 PNI68780.1 AK057266 AK093446 AK292186 AL160165 AL445645 CH471090 BC034776 BC050576 AEYP01042405 AEYP01042406 AEYP01042407 AEYP01042408 AEYP01042409 AEYP01042410 AEYP01042411 GAMS01006604 GAMP01003350 JAB16532.1 JAB49405.1 JU323001 AFE66757.1 AHZZ02007974 AQIA01023155 AQIA01023156 AQIA01023157 AQIA01023158 AQIA01023159 CM001290 EHH57017.1 JSUE03013934 CM001267 EHH23721.1 AQIB01140538 AQIB01140539 AQIB01140540 AQIB01140541 AQIB01140542 AQIB01140543 AQIB01140544 AQIB01140545 AB179151 AANG04002451 BC095649 KB030979 ELK07315.1 ADON01031071 ADON01031072 ADON01031073 BX276128 HAEB01010306 SBQ56833.1 AGTO01010822 AGTP01020290 AGTP01020291 AGTP01020292 BX510918 REGN01005372 RNA13587.1 GBSH01000210 JAG68814.1 BC079446 BFAA01000555 GCB72946.1 AL731851 AL732526 BC100481 BC109352 MCFG01000027 ORX85975.1 AADN05000150
Proteomes
UP000005204
UP000053268
UP000283053
UP000053240
UP000218220
UP000037510
+ More
UP000007266 UP000053825 UP000242457 UP000005203 UP000078542 UP000078541 UP000078492 UP000075809 UP000076502 UP000030742 UP000019118 UP000078540 UP000053097 UP000007819 UP000215335 UP000291022 UP000286642 UP000245340 UP000001595 UP000286640 UP000008912 UP000286641 UP000002254 UP000248484 UP000005225 UP000248481 UP000233200 UP000233180 UP000002277 UP000240080 UP000005640 UP000000715 UP000233160 UP000233220 UP000008225 UP000233120 UP000233080 UP000028761 UP000233060 UP000009130 UP000233100 UP000006718 UP000029965 UP000002280 UP000248482 UP000079721 UP000011712 UP000000437 UP000010552 UP000016666 UP000248480 UP000016665 UP000005215 UP000276133 UP000233020 UP000261540 UP000002494 UP000009136 UP000291021 UP000288216 UP000000589 UP000193944 UP000000539
UP000007266 UP000053825 UP000242457 UP000005203 UP000078542 UP000078541 UP000078492 UP000075809 UP000076502 UP000030742 UP000019118 UP000078540 UP000053097 UP000007819 UP000215335 UP000291022 UP000286642 UP000245340 UP000001595 UP000286640 UP000008912 UP000286641 UP000002254 UP000248484 UP000005225 UP000248481 UP000233200 UP000233180 UP000002277 UP000240080 UP000005640 UP000000715 UP000233160 UP000233220 UP000008225 UP000233120 UP000233080 UP000028761 UP000233060 UP000009130 UP000233100 UP000006718 UP000029965 UP000002280 UP000248482 UP000079721 UP000011712 UP000000437 UP000010552 UP000016666 UP000248480 UP000016665 UP000005215 UP000276133 UP000233020 UP000261540 UP000002494 UP000009136 UP000291021 UP000288216 UP000000589 UP000193944 UP000000539
Interpro
ProteinModelPortal
H9JDN6
A0A194PPY0
A0A437BXM4
A0A0N0PD18
A0A2A4K7V9
A0A0L7L2F2
+ More
A0A139WMM8 E9J5Q4 A0A0L7R566 A0A2A3EFP1 A0A088AK29 A0A195CI11 A0A310SVP4 A0A195F0M6 A0A195E742 A0A151XBE6 A0A154PI47 A0A1B6CVM8 U4UKA9 N6TH76 A0A151I186 A0A1B6L896 A0A026VVQ7 J9LPR0 A0A1B6FAH0 A0A0A9Y5J0 A0A224XAJ8 A0A232EIA6 A0A2S2NR53 A0A452SIU4 A0A3Q7Y1A4 A0A2U3VDI1 H2PTS5 A0A3Q7TVT5 G1LBZ8 A0A3Q7RUD3 U6CW53 E2QSP0 A0A2Y9EH63 H0Y2C7 A0A2Y9H6C9 A0A3P4QBT3 A0A2J8UJ55 A0A2K6NBZ3 A0A2K6KU67 H2QY34 A0A2R9B4R4 A0A2J8NAJ2 Q96MA6 M3YM49 A0A2K6FVR1 A0A2K6TA67 U3CCA3 A0A2K6E6S5 A0A2K5JYB2 H9EWJ8 A0A096P0D8 A0A2K5M076 G7PRA1 F6YCP3 A0A0D9RR30 F6PLL4 A0A2Y9J8U8 Q4R3W4 A0A1S3A1N3 A0A2I2V3K2 Q502L7 L5K726 U3I3A1 A0A2Y9DJY9 A2BFD3 A0A1A8FDG0 U3JFI3 I3M2A2 A0A2R8Q451 A0A3M7QQ56 A0A2K5D2I4 A0A3B3RYF1 A0A0B8RRT5 Q68FP8 E1BLG0 A0A452UP54 A0A401PII7 Q32M07 A0A1Y1XJS7 A0A1D5PVQ4
A0A139WMM8 E9J5Q4 A0A0L7R566 A0A2A3EFP1 A0A088AK29 A0A195CI11 A0A310SVP4 A0A195F0M6 A0A195E742 A0A151XBE6 A0A154PI47 A0A1B6CVM8 U4UKA9 N6TH76 A0A151I186 A0A1B6L896 A0A026VVQ7 J9LPR0 A0A1B6FAH0 A0A0A9Y5J0 A0A224XAJ8 A0A232EIA6 A0A2S2NR53 A0A452SIU4 A0A3Q7Y1A4 A0A2U3VDI1 H2PTS5 A0A3Q7TVT5 G1LBZ8 A0A3Q7RUD3 U6CW53 E2QSP0 A0A2Y9EH63 H0Y2C7 A0A2Y9H6C9 A0A3P4QBT3 A0A2J8UJ55 A0A2K6NBZ3 A0A2K6KU67 H2QY34 A0A2R9B4R4 A0A2J8NAJ2 Q96MA6 M3YM49 A0A2K6FVR1 A0A2K6TA67 U3CCA3 A0A2K6E6S5 A0A2K5JYB2 H9EWJ8 A0A096P0D8 A0A2K5M076 G7PRA1 F6YCP3 A0A0D9RR30 F6PLL4 A0A2Y9J8U8 Q4R3W4 A0A1S3A1N3 A0A2I2V3K2 Q502L7 L5K726 U3I3A1 A0A2Y9DJY9 A2BFD3 A0A1A8FDG0 U3JFI3 I3M2A2 A0A2R8Q451 A0A3M7QQ56 A0A2K5D2I4 A0A3B3RYF1 A0A0B8RRT5 Q68FP8 E1BLG0 A0A452UP54 A0A401PII7 Q32M07 A0A1Y1XJS7 A0A1D5PVQ4
PDB
2AK2
E-value=3.47334e-07,
Score=130
Ontologies
KEGG
PATHWAY
GO
PANTHER
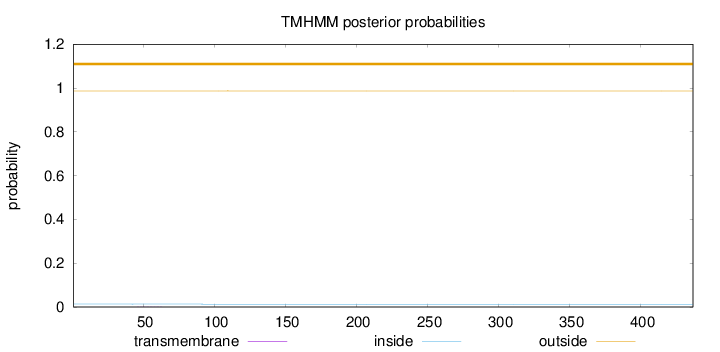
Topology
Subcellular location
Length:
437
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00751999999999998
Exp number, first 60 AAs:
0.00218
Total prob of N-in:
0.01336
outside
1 - 437
Population Genetic Test Statistics
Pi
158.581716
Theta
159.124672
Tajima's D
0.946339
CLR
0.747713
CSRT
0.644117794110294
Interpretation
Uncertain
