Pre Gene Modal
BGIBMGA007631
Annotation
PREDICTED:_nucleolar_complex_protein_4_homolog_B_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.017 Nuclear Reliability : 1.618
Sequence
CDS
ATGGCTGCTGTGCAACAAAATTTTTCGAAAATGATTTCGGGACAGCTTCGTAATAAAGCAAATGAGTTTCTTAACTCCAGAAAACATGCGAACAATCTTGCTGATATATTGCAGATGTTTGAGGTCGAAACAGACAATTATACGCCGCTAATGCTTACAATAGAAGTAATATTCACCGAACTATTAAAACGGGGCGATTTAGTCCAGGAAATAGTTCCATTAAAGCCTGCAGAACAAAGTCCAGAAGCGGAGTACACAAGATGGCTTCGTGAATGCTACGAGACTGCTCTCGCTCGTATATTAGCTTGTGTAATGAAAGGACGCACAAATTCTCGGCTGCAAGCTCTTGTCACAGCCTCCAAATTACTGCAAGTTGAAGGGAAACACCCACTGGAGCAGAGCTCTGGATATTACTTCCCATCGTTCAGATTGAAGAACATTTTCACAGTACTGTTGGACTCTGATATGTCCATGTCAGCTCCATTAGCACGTTTCCAAGAGTTCACAGAGTACAAAGATGTGCAGCAGTATGGACTGAAAGTTCTCTCTACCCTGGCATATCGGAAGTCTCCCACTACAGTCTACATGCAGAATTACTTGGAACTTCTTGACAAACTACTAATCTCAGAGATTGCCACTGAGACAAAAACTAAAAATAACAAAGGTGGTGATGACCATGAAGAAGAATTTAAAGTTCTATGTGGCACAAATGCCAAGGTGCCATTCCCGTACAACCCGGGCGTGTGTCGTCGCTACGCCAACCGCTGCTGGAGCTTCGCGTGCCAGTGGCCGCTGTGCTCGGAGCCGCGCACGCACCGCCGCGCCCTGCTGCTGCTGGTGGAGCGCCTCATGCCGCTGCTCAGCAAGCCGCACCTCGCCACCGACATGCTCTGCGACAGTCTCGACGCCGGCGGTCCGATCAGCATGCTGGCGCTGCAGGGCGTGCTGGAGCTGGTGCGGCGGCACAACGTCGACTACCCGGACATGTACGACCGCCTGTACGCGATGTTCGAGCCGGAGATGTTCGCCACGCGCTACAAGCGGCGCCTCATGCACCTCGCCGACATCTTCCTCAGCTCCACACATTTGCCTGAGAGTTTGGTGGCGGCCTTCGCAAAGCGTCTGTCTCGGCTGGCGTTGGTGGCCTGCCCGGAGGACGCCGTGGGGCTGCTGCAGCTCATAGCCAACCTGCTGCTCCGACATCCGGCGTTGAAGCGCATGATCTGCTACGAAGATACGCCCGCCATCATGTCGAGCGACCCGTACGTGATGGAGGAGACGTGCGCGGCGCGCGCGGGCGCGCTGGGCTCGTCGCTGTGGGAGGTGGCCGCGCTGCGCCGCCACGCCGCGCCCGCGCTCTCCGCCGCGCTCGCCGCCGTGCTCGCCGCCGCCGACCCGCGCCCCGAGCCCTTCAACCTGTCCTCCGGGGACGCCAAGATGTTTGATGCTGAGTTGAAGAAAAGATTCAAAACGATCGAGATGAACTTTATCCGGCCTCAAGGTATGACTGCGCCCTCTGGCGAACGTCTCATGCAGTACTGGGAACTGATGGCTTGA
Protein
MAAVQQNFSKMISGQLRNKANEFLNSRKHANNLADILQMFEVETDNYTPLMLTIEVIFTELLKRGDLVQEIVPLKPAEQSPEAEYTRWLRECYETALARILACVMKGRTNSRLQALVTASKLLQVEGKHPLEQSSGYYFPSFRLKNIFTVLLDSDMSMSAPLARFQEFTEYKDVQQYGLKVLSTLAYRKSPTTVYMQNYLELLDKLLISEIATETKTKNNKGGDDHEEEFKVLCGTNAKVPFPYNPGVCRRYANRCWSFACQWPLCSEPRTHRRALLLLVERLMPLLSKPHLATDMLCDSLDAGGPISMLALQGVLELVRRHNVDYPDMYDRLYAMFEPEMFATRYKRRLMHLADIFLSSTHLPESLVAAFAKRLSRLALVACPEDAVGLLQLIANLLLRHPALKRMICYEDTPAIMSSDPYVMEETCAARAGALGSSLWEVAALRRHAAPALSAALAAVLAAADPRPEPFNLSSGDAKMFDAELKKRFKTIEMNFIRPQGMTAPSGERLMQYWELMA
Summary
Uniprot
H9JDN4
A0A2H1WWW0
A0A194PPJ7
A0A212FB82
A0A3S2PLW8
A0A0N1PJE0
+ More
A0A0L7R5D7 W8CCQ3 E2BBR0 A0A2A3EHG3 A0A0J7L4F7 A0A088ACF1 E9J3P6 A0A154PJ69 A0A0A1X3T4 A0A026WAS5 F4W673 A0A195AVM6 A0A195DMR0 A0A158P202 A0A1L8DRN8 A0A0N0U6Z2 A0A0K8TKR7 A0A034WCQ9 E2AY71 A0A0K8WB17 A0A151IKK9 A0A195FJ64 A0A067R6T6 A0A1L8DRT8 A0A0C9R7I8 A0A2P8XU77 A0A0T6B489 A0A232F4Z6 A0A0L0C3S7 A0A1W4WU83 A0A1B0CBF0 A0A1Y1L8R3 A0A1I8QA03 A0A182H461 Q17FJ6 A0A182G892 A0A023ETC8 A0A1A9VNE4 A0A1B0AFF0 D3TLC4 A0A084VRU3 A0A1A9WA77 A0A1B0AUC8 D6X118 U5EV79 A0A1Q3F4U9 U4UIX0 A0A1A9XKG1 A0A182J377 Q1DGD8 A0A1B0FQ44 B0XHP9 A0A1J1HJW1 A0A336LY15 A0A1B6E3T7 A0A336KXB3 A0A1I8M756 A0A182LTX1 A0A182QP55 A0A2M4AD98 A0A2M4AD79 A0A182SIP4 Q7PYC7 W5J6F2 A0A2C9GRN0 N6SSA2 A0A2M4BK23 A0A2M4AF38 A0A2M4ADI1 A0A2M3Z4X7 A0A2M3Z4Y5 A0A2M4BK47 T1DQF0 A0A182KM64 A0A182YJW0 T1DQF7 A0A182UTN7 A0A182R1H1 A0A182VQW2 A0A182N4W6 A0A182XBN4 A0A182UJU0 A0A182K4S0 A0A1Q3F522 A0A182PEE2 V5GYM6 A0A2J7R095 A0A1S3DTP4 B4L494
A0A0L7R5D7 W8CCQ3 E2BBR0 A0A2A3EHG3 A0A0J7L4F7 A0A088ACF1 E9J3P6 A0A154PJ69 A0A0A1X3T4 A0A026WAS5 F4W673 A0A195AVM6 A0A195DMR0 A0A158P202 A0A1L8DRN8 A0A0N0U6Z2 A0A0K8TKR7 A0A034WCQ9 E2AY71 A0A0K8WB17 A0A151IKK9 A0A195FJ64 A0A067R6T6 A0A1L8DRT8 A0A0C9R7I8 A0A2P8XU77 A0A0T6B489 A0A232F4Z6 A0A0L0C3S7 A0A1W4WU83 A0A1B0CBF0 A0A1Y1L8R3 A0A1I8QA03 A0A182H461 Q17FJ6 A0A182G892 A0A023ETC8 A0A1A9VNE4 A0A1B0AFF0 D3TLC4 A0A084VRU3 A0A1A9WA77 A0A1B0AUC8 D6X118 U5EV79 A0A1Q3F4U9 U4UIX0 A0A1A9XKG1 A0A182J377 Q1DGD8 A0A1B0FQ44 B0XHP9 A0A1J1HJW1 A0A336LY15 A0A1B6E3T7 A0A336KXB3 A0A1I8M756 A0A182LTX1 A0A182QP55 A0A2M4AD98 A0A2M4AD79 A0A182SIP4 Q7PYC7 W5J6F2 A0A2C9GRN0 N6SSA2 A0A2M4BK23 A0A2M4AF38 A0A2M4ADI1 A0A2M3Z4X7 A0A2M3Z4Y5 A0A2M4BK47 T1DQF0 A0A182KM64 A0A182YJW0 T1DQF7 A0A182UTN7 A0A182R1H1 A0A182VQW2 A0A182N4W6 A0A182XBN4 A0A182UJU0 A0A182K4S0 A0A1Q3F522 A0A182PEE2 V5GYM6 A0A2J7R095 A0A1S3DTP4 B4L494
Pubmed
19121390
26354079
22118469
24495485
20798317
21282665
+ More
25830018 24508170 30249741 21719571 21347285 26369729 25348373 24845553 29403074 28648823 26108605 28004739 26483478 17510324 24945155 20353571 24438588 18362917 19820115 23537049 25315136 12364791 14747013 17210077 20920257 23761445 20966253 25244985 17994087
25830018 24508170 30249741 21719571 21347285 26369729 25348373 24845553 29403074 28648823 26108605 28004739 26483478 17510324 24945155 20353571 24438588 18362917 19820115 23537049 25315136 12364791 14747013 17210077 20920257 23761445 20966253 25244985 17994087
EMBL
BABH01003239
ODYU01011611
SOQ57456.1
KQ459596
KPI95366.1
AGBW02009363
+ More
OWR51002.1 RSAL01000001 RVE55218.1 KQ460497 KPJ14324.1 KQ414654 KOC66034.1 GAMC01003296 JAC03260.1 GL447151 EFN86836.1 KZ288256 PBC30616.1 LBMM01000815 KMQ97428.1 GL768069 EFZ12458.1 KQ434923 KZC11504.1 GBXI01008999 JAD05293.1 KK107293 QOIP01000005 EZA53150.1 RLU23110.1 GL887707 EGI70208.1 KQ976731 KYM76293.1 KQ980724 KYN14117.1 ADTU01006919 GFDF01004962 JAV09122.1 KQ435712 KOX79371.1 GDAI01002636 JAI14967.1 GAKP01006850 JAC52102.1 GL443762 EFN61606.1 GDHF01003926 JAI48388.1 KQ977199 KYN05000.1 KQ981523 KYN40448.1 KK852703 KDR18092.1 GFDF01005000 JAV09084.1 GBYB01003990 JAG73757.1 PYGN01001338 PSN35555.1 LJIG01016005 KRT81901.1 NNAY01000998 OXU25550.1 JRES01000948 KNC26907.1 AJWK01005094 AJWK01005095 GEZM01064541 JAV68730.1 JXUM01108816 JXUM01108817 JXUM01108818 JXUM01108819 JXUM01108820 JXUM01108821 KQ565266 KXJ71153.1 CH477270 EAT45347.1 JXUM01151632 KQ571263 KXJ68214.1 GAPW01001008 JAC12590.1 EZ422226 ADD18502.1 ATLV01015780 KE525036 KFB40687.1 JXJN01003606 KQ971367 EFA09396.1 GANO01001980 JAB57891.1 GFDL01012461 JAV22584.1 KB632343 ERL93022.1 CH902499 EAT32217.1 CCAG010006700 DS233189 EDS28556.1 CVRI01000002 CRK86758.1 UFQS01000218 UFQT01000218 SSX01501.1 SSX21881.1 GEDC01004741 JAS32557.1 UFQS01001264 UFQT01001264 SSX10041.1 SSX29763.1 AXCM01002674 AXCN02000187 GGFK01005409 MBW38730.1 GGFK01005389 MBW38710.1 AAAB01008987 EAA01545.4 EGK97452.1 ADMH02002134 ETN58410.1 APCN01002077 APGK01058607 KB741291 ENN70524.1 GGFJ01004288 MBW53429.1 GGFK01006073 MBW39394.1 GGFK01005461 MBW38782.1 GGFM01002815 MBW23566.1 GGFM01002840 MBW23591.1 GGFJ01004289 MBW53430.1 GAMD01002559 JAA99031.1 GAMD01002529 JAA99061.1 GFDL01012390 JAV22655.1 GALX01001504 JAB66962.1 NEVH01008283 PNF34244.1 CH933810 EDW07372.1
OWR51002.1 RSAL01000001 RVE55218.1 KQ460497 KPJ14324.1 KQ414654 KOC66034.1 GAMC01003296 JAC03260.1 GL447151 EFN86836.1 KZ288256 PBC30616.1 LBMM01000815 KMQ97428.1 GL768069 EFZ12458.1 KQ434923 KZC11504.1 GBXI01008999 JAD05293.1 KK107293 QOIP01000005 EZA53150.1 RLU23110.1 GL887707 EGI70208.1 KQ976731 KYM76293.1 KQ980724 KYN14117.1 ADTU01006919 GFDF01004962 JAV09122.1 KQ435712 KOX79371.1 GDAI01002636 JAI14967.1 GAKP01006850 JAC52102.1 GL443762 EFN61606.1 GDHF01003926 JAI48388.1 KQ977199 KYN05000.1 KQ981523 KYN40448.1 KK852703 KDR18092.1 GFDF01005000 JAV09084.1 GBYB01003990 JAG73757.1 PYGN01001338 PSN35555.1 LJIG01016005 KRT81901.1 NNAY01000998 OXU25550.1 JRES01000948 KNC26907.1 AJWK01005094 AJWK01005095 GEZM01064541 JAV68730.1 JXUM01108816 JXUM01108817 JXUM01108818 JXUM01108819 JXUM01108820 JXUM01108821 KQ565266 KXJ71153.1 CH477270 EAT45347.1 JXUM01151632 KQ571263 KXJ68214.1 GAPW01001008 JAC12590.1 EZ422226 ADD18502.1 ATLV01015780 KE525036 KFB40687.1 JXJN01003606 KQ971367 EFA09396.1 GANO01001980 JAB57891.1 GFDL01012461 JAV22584.1 KB632343 ERL93022.1 CH902499 EAT32217.1 CCAG010006700 DS233189 EDS28556.1 CVRI01000002 CRK86758.1 UFQS01000218 UFQT01000218 SSX01501.1 SSX21881.1 GEDC01004741 JAS32557.1 UFQS01001264 UFQT01001264 SSX10041.1 SSX29763.1 AXCM01002674 AXCN02000187 GGFK01005409 MBW38730.1 GGFK01005389 MBW38710.1 AAAB01008987 EAA01545.4 EGK97452.1 ADMH02002134 ETN58410.1 APCN01002077 APGK01058607 KB741291 ENN70524.1 GGFJ01004288 MBW53429.1 GGFK01006073 MBW39394.1 GGFK01005461 MBW38782.1 GGFM01002815 MBW23566.1 GGFM01002840 MBW23591.1 GGFJ01004289 MBW53430.1 GAMD01002559 JAA99031.1 GAMD01002529 JAA99061.1 GFDL01012390 JAV22655.1 GALX01001504 JAB66962.1 NEVH01008283 PNF34244.1 CH933810 EDW07372.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000283053
UP000053240
UP000053825
+ More
UP000008237 UP000242457 UP000036403 UP000005203 UP000076502 UP000053097 UP000279307 UP000007755 UP000078540 UP000078492 UP000005205 UP000053105 UP000000311 UP000078542 UP000078541 UP000027135 UP000245037 UP000215335 UP000037069 UP000192223 UP000092461 UP000095300 UP000069940 UP000249989 UP000008820 UP000078200 UP000092445 UP000030765 UP000091820 UP000092460 UP000007266 UP000030742 UP000092443 UP000075880 UP000092444 UP000002320 UP000183832 UP000095301 UP000075883 UP000075886 UP000075901 UP000007062 UP000000673 UP000075840 UP000019118 UP000075882 UP000076408 UP000075903 UP000075900 UP000075920 UP000075884 UP000076407 UP000075902 UP000075881 UP000075885 UP000235965 UP000079169 UP000009192
UP000008237 UP000242457 UP000036403 UP000005203 UP000076502 UP000053097 UP000279307 UP000007755 UP000078540 UP000078492 UP000005205 UP000053105 UP000000311 UP000078542 UP000078541 UP000027135 UP000245037 UP000215335 UP000037069 UP000192223 UP000092461 UP000095300 UP000069940 UP000249989 UP000008820 UP000078200 UP000092445 UP000030765 UP000091820 UP000092460 UP000007266 UP000030742 UP000092443 UP000075880 UP000092444 UP000002320 UP000183832 UP000095301 UP000075883 UP000075886 UP000075901 UP000007062 UP000000673 UP000075840 UP000019118 UP000075882 UP000076408 UP000075903 UP000075900 UP000075920 UP000075884 UP000076407 UP000075902 UP000075881 UP000075885 UP000235965 UP000079169 UP000009192
PRIDE
Interpro
IPR027193
Noc4
+ More
IPR005612 CCAAT-binding_factor
IPR001251 CRAL-TRIO_dom
IPR019775 WD40_repeat_CS
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR002293 AA/rel_permease1
IPR027973 DUF4602
IPR008858 TROVE_dom
IPR037214 TROVE_dom_sf
IPR036465 vWFA_dom_sf
IPR040322 TROVE2
IPR016024 ARM-type_fold
IPR005612 CCAAT-binding_factor
IPR001251 CRAL-TRIO_dom
IPR019775 WD40_repeat_CS
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR017986 WD40_repeat_dom
IPR002293 AA/rel_permease1
IPR027973 DUF4602
IPR008858 TROVE_dom
IPR037214 TROVE_dom_sf
IPR036465 vWFA_dom_sf
IPR040322 TROVE2
IPR016024 ARM-type_fold
Gene 3D
ProteinModelPortal
H9JDN4
A0A2H1WWW0
A0A194PPJ7
A0A212FB82
A0A3S2PLW8
A0A0N1PJE0
+ More
A0A0L7R5D7 W8CCQ3 E2BBR0 A0A2A3EHG3 A0A0J7L4F7 A0A088ACF1 E9J3P6 A0A154PJ69 A0A0A1X3T4 A0A026WAS5 F4W673 A0A195AVM6 A0A195DMR0 A0A158P202 A0A1L8DRN8 A0A0N0U6Z2 A0A0K8TKR7 A0A034WCQ9 E2AY71 A0A0K8WB17 A0A151IKK9 A0A195FJ64 A0A067R6T6 A0A1L8DRT8 A0A0C9R7I8 A0A2P8XU77 A0A0T6B489 A0A232F4Z6 A0A0L0C3S7 A0A1W4WU83 A0A1B0CBF0 A0A1Y1L8R3 A0A1I8QA03 A0A182H461 Q17FJ6 A0A182G892 A0A023ETC8 A0A1A9VNE4 A0A1B0AFF0 D3TLC4 A0A084VRU3 A0A1A9WA77 A0A1B0AUC8 D6X118 U5EV79 A0A1Q3F4U9 U4UIX0 A0A1A9XKG1 A0A182J377 Q1DGD8 A0A1B0FQ44 B0XHP9 A0A1J1HJW1 A0A336LY15 A0A1B6E3T7 A0A336KXB3 A0A1I8M756 A0A182LTX1 A0A182QP55 A0A2M4AD98 A0A2M4AD79 A0A182SIP4 Q7PYC7 W5J6F2 A0A2C9GRN0 N6SSA2 A0A2M4BK23 A0A2M4AF38 A0A2M4ADI1 A0A2M3Z4X7 A0A2M3Z4Y5 A0A2M4BK47 T1DQF0 A0A182KM64 A0A182YJW0 T1DQF7 A0A182UTN7 A0A182R1H1 A0A182VQW2 A0A182N4W6 A0A182XBN4 A0A182UJU0 A0A182K4S0 A0A1Q3F522 A0A182PEE2 V5GYM6 A0A2J7R095 A0A1S3DTP4 B4L494
A0A0L7R5D7 W8CCQ3 E2BBR0 A0A2A3EHG3 A0A0J7L4F7 A0A088ACF1 E9J3P6 A0A154PJ69 A0A0A1X3T4 A0A026WAS5 F4W673 A0A195AVM6 A0A195DMR0 A0A158P202 A0A1L8DRN8 A0A0N0U6Z2 A0A0K8TKR7 A0A034WCQ9 E2AY71 A0A0K8WB17 A0A151IKK9 A0A195FJ64 A0A067R6T6 A0A1L8DRT8 A0A0C9R7I8 A0A2P8XU77 A0A0T6B489 A0A232F4Z6 A0A0L0C3S7 A0A1W4WU83 A0A1B0CBF0 A0A1Y1L8R3 A0A1I8QA03 A0A182H461 Q17FJ6 A0A182G892 A0A023ETC8 A0A1A9VNE4 A0A1B0AFF0 D3TLC4 A0A084VRU3 A0A1A9WA77 A0A1B0AUC8 D6X118 U5EV79 A0A1Q3F4U9 U4UIX0 A0A1A9XKG1 A0A182J377 Q1DGD8 A0A1B0FQ44 B0XHP9 A0A1J1HJW1 A0A336LY15 A0A1B6E3T7 A0A336KXB3 A0A1I8M756 A0A182LTX1 A0A182QP55 A0A2M4AD98 A0A2M4AD79 A0A182SIP4 Q7PYC7 W5J6F2 A0A2C9GRN0 N6SSA2 A0A2M4BK23 A0A2M4AF38 A0A2M4ADI1 A0A2M3Z4X7 A0A2M3Z4Y5 A0A2M4BK47 T1DQF0 A0A182KM64 A0A182YJW0 T1DQF7 A0A182UTN7 A0A182R1H1 A0A182VQW2 A0A182N4W6 A0A182XBN4 A0A182UJU0 A0A182K4S0 A0A1Q3F522 A0A182PEE2 V5GYM6 A0A2J7R095 A0A1S3DTP4 B4L494
PDB
5WLC
E-value=2.26573e-21,
Score=253
Ontologies
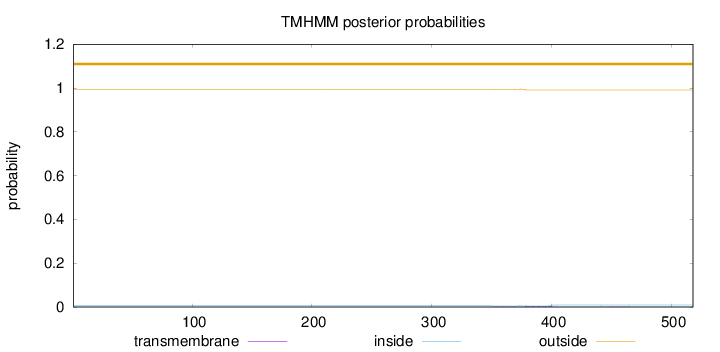
Topology
Length:
518
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13487
Exp number, first 60 AAs:
0.00093
Total prob of N-in:
0.00555
outside
1 - 518
Population Genetic Test Statistics
Pi
208.611432
Theta
173.390742
Tajima's D
1.091123
CLR
0.165701
CSRT
0.685915704214789
Interpretation
Uncertain
