Pre Gene Modal
BGIBMGA007955
Annotation
hypothetical_protein_KGM_01538_[Danaus_plexippus]
Full name
Probable ribosome production factor 1
Alternative Name
Ribosome biogenesis protein RPF1
Location in the cell
Nuclear Reliability : 4.32
Sequence
CDS
ATGGCTCGTGGAAAGGATAAAAAAAAACGCAAGTTAGAAAAGAAACAAGAGGGAGATGATGTACCTAAGAAAATCCCTCACACGTTGGAAACTCTTCGTGAAAAAGATGAGACGATGCTCGTTAATGCTGACACCGAGGAACAAGAGGAGGCACAAAAAGATATGGAACTTGATGAACTTTCAACATACTATGAGAATTCGTATGAGCCAAAAGTGCTGATCACATACTCTGATAATCCACACAGTAAAACTAGAATATTCGGAAGAGAACTGACTAGGATTATACCTAATTCTTTATCGAGATACAGACAGAGGTCATCAGTGAAACGTATAGTACAATCAGCAATCAGAGAAGACGTCACAGATGTGATTATAATTAATGAGAACCAACGGCAGCCTAATGGTTTTCTTCTCATCCATTTGCCTGCAGGTCCCACTGCGCACTTCCGTCTTTCTAGCTACCAGAGGTCGTCCTCAACAACTTCAGTACACGATTGGGTCTGA
Protein
MARGKDKKKRKLEKKQEGDDVPKKIPHTLETLREKDETMLVNADTEEQEEAQKDMELDELSTYYENSYEPKVLITYSDNPHSKTRIFGRELTRIIPNSLSRYRQRSSVKRIVQSAIREDVTDVIIINENQRQPNGFLLIHLPAGPTAHFRLSSYQRSSSTTSVHDWV
Summary
Description
May be required for ribosome biogenesis.
Keywords
Complete proteome
Nucleus
Reference proteome
Ribosome biogenesis
RNA-binding
rRNA processing
rRNA-binding
Feature
chain Probable ribosome production factor 1
Uniprot
H9JEK8
A0A1E1VZR5
A0A212EYL4
A0A3S2MAL1
A0A2H1WYJ3
A0A2A4K161
+ More
A0A194PQX6 A0A0N0PD15 S4P272 A0A2J7RG04 A0A2J7RG25 V5GF00 F4WHG9 A0A2J7RG17 A0A158N933 A0A1Y1KTM5 A0A195FU19 A0A0N0BFX7 A0A067QIZ0 A0A1W4WU45 D6WGH3 E2A6R2 A0A151X4Q2 A0A0J7K897 A0A1J1ISH1 A0A151IQ65 A0A232EV81 K7ITH8 A0A026VY58 A0A088ALI3 E2BSA6 A0A195BUV9 A0A0L7QMJ7 A0A151J863 A0A1I8PMP0 A0A336L1R1 A0A154P2V0 A0A1I8MCX0 A0A1W4VB28 T1PAI8 B4Q3Q1 A0A2A3E8P1 W8BS10 B3MVH7 B4P1Y5 B4IE63 D3TPY9 A0A1A9WKW3 A0A1B0CSC9 B3N3X3 Q9VKB4 A0A1B0FBV4 A0A034WHK6 A0A034WFZ9 A0A1A9UL16 A0A3B0K0U3 A0A0K8VYJ9 A0A0A1WYA7 A0A1A9XCS3 A0A1B0C454 B4N772 A0A1B6KQC3 N6UM87 A0A1L8DG24 Q29NK6 A0A170YNZ4 B4GQK3 A0A1B0D141 A0A069DR55 A0A0L0C6C4 B4KI01 A0A1Q3F1M4 A0A0M4E696 A0A1B6EXZ7 A0A1L8DG36 B4LSF5 B0WPW6 B4JBL6 A0A0A9WF81 A0A224XTC4 A0A1S4F9I7 A0A0V0G8E4 Q17BF6 A0A1B6JQ02 A0A2M4AP89 A0A2M4AKL6 A0A2M4AKI6 A0A182F2A7 A0A2M3Z8G6 A0A2M4BRC4 A0A2M3Z7I5 A0A2M4BRD0 W5J427 A0A2M4CQI9 A0A0T6BD34 A0A182RWS9 A0A0P4VMU7 E0VHX2
A0A194PQX6 A0A0N0PD15 S4P272 A0A2J7RG04 A0A2J7RG25 V5GF00 F4WHG9 A0A2J7RG17 A0A158N933 A0A1Y1KTM5 A0A195FU19 A0A0N0BFX7 A0A067QIZ0 A0A1W4WU45 D6WGH3 E2A6R2 A0A151X4Q2 A0A0J7K897 A0A1J1ISH1 A0A151IQ65 A0A232EV81 K7ITH8 A0A026VY58 A0A088ALI3 E2BSA6 A0A195BUV9 A0A0L7QMJ7 A0A151J863 A0A1I8PMP0 A0A336L1R1 A0A154P2V0 A0A1I8MCX0 A0A1W4VB28 T1PAI8 B4Q3Q1 A0A2A3E8P1 W8BS10 B3MVH7 B4P1Y5 B4IE63 D3TPY9 A0A1A9WKW3 A0A1B0CSC9 B3N3X3 Q9VKB4 A0A1B0FBV4 A0A034WHK6 A0A034WFZ9 A0A1A9UL16 A0A3B0K0U3 A0A0K8VYJ9 A0A0A1WYA7 A0A1A9XCS3 A0A1B0C454 B4N772 A0A1B6KQC3 N6UM87 A0A1L8DG24 Q29NK6 A0A170YNZ4 B4GQK3 A0A1B0D141 A0A069DR55 A0A0L0C6C4 B4KI01 A0A1Q3F1M4 A0A0M4E696 A0A1B6EXZ7 A0A1L8DG36 B4LSF5 B0WPW6 B4JBL6 A0A0A9WF81 A0A224XTC4 A0A1S4F9I7 A0A0V0G8E4 Q17BF6 A0A1B6JQ02 A0A2M4AP89 A0A2M4AKL6 A0A2M4AKI6 A0A182F2A7 A0A2M3Z8G6 A0A2M4BRC4 A0A2M3Z7I5 A0A2M4BRD0 W5J427 A0A2M4CQI9 A0A0T6BD34 A0A182RWS9 A0A0P4VMU7 E0VHX2
Pubmed
19121390
22118469
26354079
23622113
21719571
21347285
+ More
28004739 24845553 18362917 19820115 20798317 28648823 20075255 24508170 30249741 25315136 17994087 22936249 24495485 17550304 20353571 10731132 12537572 12537569 25348373 25830018 23537049 15632085 26334808 26108605 25401762 26823975 17510324 20920257 23761445 27129103 20566863
28004739 24845553 18362917 19820115 20798317 28648823 20075255 24508170 30249741 25315136 17994087 22936249 24495485 17550304 20353571 10731132 12537572 12537569 25348373 25830018 23537049 15632085 26334808 26108605 25401762 26823975 17510324 20920257 23761445 27129103 20566863
EMBL
BABH01003240
BABH01003241
GDQN01010903
JAT80151.1
AGBW02011483
OWR46575.1
+ More
RSAL01000001 RVE55220.1 ODYU01011611 SOQ57454.1 NWSH01000281 PCG77799.1 KQ459596 KPI95363.1 KQ460497 KPJ14320.1 GAIX01012165 JAA80395.1 NEVH01004406 PNF39769.1 PNF39770.1 GALX01008204 JAB60262.1 GL888161 EGI66323.1 PNF39768.1 ADTU01009059 GEZM01075835 JAV63928.1 KQ981268 KYN43921.1 KQ435794 KOX73741.1 KK853463 KDR07446.1 KQ971321 EFA00559.1 GL437184 EFN70879.1 KQ982543 KYQ55357.1 LBMM01012056 KMQ86441.1 CVRI01000055 CRL01473.1 KQ976785 KYN08304.1 NNAY01002042 OXU22245.1 AAZX01003331 KK107638 QOIP01000004 EZA48406.1 RLU23505.1 GL450157 EFN81419.1 KQ976405 KYM91865.1 KQ414889 KOC59843.1 KQ979640 KYN20047.1 UFQS01001365 UFQT01001365 SSX10529.1 SSX30215.1 KQ434807 KZC06153.1 KA645170 AFP59799.1 CM000361 CM002910 EDX04776.1 KMY89875.1 KZ288324 PBC28087.1 GAMC01002400 JAC04156.1 CH902624 EDV33242.1 CM000157 EDW88156.1 CH480831 EDW45890.1 EZ423491 ADD19767.1 AJWK01008028 AJWK01025908 CH954177 EDV58825.1 AE014134 AY051683 CCAG010019860 GAKP01005734 JAC53218.1 GAKP01005735 JAC53217.1 OUUW01000006 SPP81450.1 GDHF01008377 JAI43937.1 GBXI01010914 JAD03378.1 JXJN01025274 CH964182 EDW80213.1 GEBQ01026328 JAT13649.1 APGK01026508 KB740635 KB631992 ENN79827.1 ERL87766.1 GFDF01008676 JAV05408.1 CH379059 EAL34213.2 GEMB01003106 JAS00103.1 CH479187 EDW39875.1 AJVK01010182 GBGD01002336 JAC86553.1 JRES01000905 JRES01000841 KNC27370.1 KNC27830.1 CH933807 EDW11283.1 GFDL01013591 JAV21454.1 CP012523 ALC38065.1 ALC39381.1 GECZ01026952 JAS42817.1 GFDF01008675 JAV05409.1 CH940649 EDW64777.1 DS232030 EDS32554.1 CH916368 EDW04039.1 GBHO01038456 GBRD01012575 GDHC01004851 JAG05148.1 JAG53249.1 JAQ13778.1 GFTR01004654 JAW11772.1 GECL01001939 JAP04185.1 CH477322 EAT43588.1 GECU01006501 JAT01206.1 GGFK01009268 MBW42589.1 GGFK01008015 MBW41336.1 GGFK01007985 MBW41306.1 GGFM01004029 MBW24780.1 GGFJ01006496 MBW55637.1 GGFM01003699 MBW24450.1 GGFJ01006495 MBW55636.1 ADMH02002168 ETN58088.1 GGFL01003422 MBW67600.1 LJIG01001682 KRT85258.1 GDKW01002948 JAI53647.1 DS235172 EEB12978.1
RSAL01000001 RVE55220.1 ODYU01011611 SOQ57454.1 NWSH01000281 PCG77799.1 KQ459596 KPI95363.1 KQ460497 KPJ14320.1 GAIX01012165 JAA80395.1 NEVH01004406 PNF39769.1 PNF39770.1 GALX01008204 JAB60262.1 GL888161 EGI66323.1 PNF39768.1 ADTU01009059 GEZM01075835 JAV63928.1 KQ981268 KYN43921.1 KQ435794 KOX73741.1 KK853463 KDR07446.1 KQ971321 EFA00559.1 GL437184 EFN70879.1 KQ982543 KYQ55357.1 LBMM01012056 KMQ86441.1 CVRI01000055 CRL01473.1 KQ976785 KYN08304.1 NNAY01002042 OXU22245.1 AAZX01003331 KK107638 QOIP01000004 EZA48406.1 RLU23505.1 GL450157 EFN81419.1 KQ976405 KYM91865.1 KQ414889 KOC59843.1 KQ979640 KYN20047.1 UFQS01001365 UFQT01001365 SSX10529.1 SSX30215.1 KQ434807 KZC06153.1 KA645170 AFP59799.1 CM000361 CM002910 EDX04776.1 KMY89875.1 KZ288324 PBC28087.1 GAMC01002400 JAC04156.1 CH902624 EDV33242.1 CM000157 EDW88156.1 CH480831 EDW45890.1 EZ423491 ADD19767.1 AJWK01008028 AJWK01025908 CH954177 EDV58825.1 AE014134 AY051683 CCAG010019860 GAKP01005734 JAC53218.1 GAKP01005735 JAC53217.1 OUUW01000006 SPP81450.1 GDHF01008377 JAI43937.1 GBXI01010914 JAD03378.1 JXJN01025274 CH964182 EDW80213.1 GEBQ01026328 JAT13649.1 APGK01026508 KB740635 KB631992 ENN79827.1 ERL87766.1 GFDF01008676 JAV05408.1 CH379059 EAL34213.2 GEMB01003106 JAS00103.1 CH479187 EDW39875.1 AJVK01010182 GBGD01002336 JAC86553.1 JRES01000905 JRES01000841 KNC27370.1 KNC27830.1 CH933807 EDW11283.1 GFDL01013591 JAV21454.1 CP012523 ALC38065.1 ALC39381.1 GECZ01026952 JAS42817.1 GFDF01008675 JAV05409.1 CH940649 EDW64777.1 DS232030 EDS32554.1 CH916368 EDW04039.1 GBHO01038456 GBRD01012575 GDHC01004851 JAG05148.1 JAG53249.1 JAQ13778.1 GFTR01004654 JAW11772.1 GECL01001939 JAP04185.1 CH477322 EAT43588.1 GECU01006501 JAT01206.1 GGFK01009268 MBW42589.1 GGFK01008015 MBW41336.1 GGFK01007985 MBW41306.1 GGFM01004029 MBW24780.1 GGFJ01006496 MBW55637.1 GGFM01003699 MBW24450.1 GGFJ01006495 MBW55636.1 ADMH02002168 ETN58088.1 GGFL01003422 MBW67600.1 LJIG01001682 KRT85258.1 GDKW01002948 JAI53647.1 DS235172 EEB12978.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000218220
UP000053268
UP000053240
+ More
UP000235965 UP000007755 UP000005205 UP000078541 UP000053105 UP000027135 UP000192223 UP000007266 UP000000311 UP000075809 UP000036403 UP000183832 UP000078542 UP000215335 UP000002358 UP000053097 UP000279307 UP000005203 UP000008237 UP000078540 UP000053825 UP000078492 UP000095300 UP000076502 UP000095301 UP000192221 UP000000304 UP000242457 UP000007801 UP000002282 UP000001292 UP000091820 UP000092461 UP000008711 UP000000803 UP000092444 UP000078200 UP000268350 UP000092443 UP000092460 UP000007798 UP000019118 UP000030742 UP000001819 UP000008744 UP000092462 UP000037069 UP000009192 UP000092553 UP000008792 UP000002320 UP000001070 UP000008820 UP000069272 UP000000673 UP000075900 UP000009046
UP000235965 UP000007755 UP000005205 UP000078541 UP000053105 UP000027135 UP000192223 UP000007266 UP000000311 UP000075809 UP000036403 UP000183832 UP000078542 UP000215335 UP000002358 UP000053097 UP000279307 UP000005203 UP000008237 UP000078540 UP000053825 UP000078492 UP000095300 UP000076502 UP000095301 UP000192221 UP000000304 UP000242457 UP000007801 UP000002282 UP000001292 UP000091820 UP000092461 UP000008711 UP000000803 UP000092444 UP000078200 UP000268350 UP000092443 UP000092460 UP000007798 UP000019118 UP000030742 UP000001819 UP000008744 UP000092462 UP000037069 UP000009192 UP000092553 UP000008792 UP000002320 UP000001070 UP000008820 UP000069272 UP000000673 UP000075900 UP000009046
Pfam
PF04427 Brix
Interpro
IPR007109
Brix
ProteinModelPortal
H9JEK8
A0A1E1VZR5
A0A212EYL4
A0A3S2MAL1
A0A2H1WYJ3
A0A2A4K161
+ More
A0A194PQX6 A0A0N0PD15 S4P272 A0A2J7RG04 A0A2J7RG25 V5GF00 F4WHG9 A0A2J7RG17 A0A158N933 A0A1Y1KTM5 A0A195FU19 A0A0N0BFX7 A0A067QIZ0 A0A1W4WU45 D6WGH3 E2A6R2 A0A151X4Q2 A0A0J7K897 A0A1J1ISH1 A0A151IQ65 A0A232EV81 K7ITH8 A0A026VY58 A0A088ALI3 E2BSA6 A0A195BUV9 A0A0L7QMJ7 A0A151J863 A0A1I8PMP0 A0A336L1R1 A0A154P2V0 A0A1I8MCX0 A0A1W4VB28 T1PAI8 B4Q3Q1 A0A2A3E8P1 W8BS10 B3MVH7 B4P1Y5 B4IE63 D3TPY9 A0A1A9WKW3 A0A1B0CSC9 B3N3X3 Q9VKB4 A0A1B0FBV4 A0A034WHK6 A0A034WFZ9 A0A1A9UL16 A0A3B0K0U3 A0A0K8VYJ9 A0A0A1WYA7 A0A1A9XCS3 A0A1B0C454 B4N772 A0A1B6KQC3 N6UM87 A0A1L8DG24 Q29NK6 A0A170YNZ4 B4GQK3 A0A1B0D141 A0A069DR55 A0A0L0C6C4 B4KI01 A0A1Q3F1M4 A0A0M4E696 A0A1B6EXZ7 A0A1L8DG36 B4LSF5 B0WPW6 B4JBL6 A0A0A9WF81 A0A224XTC4 A0A1S4F9I7 A0A0V0G8E4 Q17BF6 A0A1B6JQ02 A0A2M4AP89 A0A2M4AKL6 A0A2M4AKI6 A0A182F2A7 A0A2M3Z8G6 A0A2M4BRC4 A0A2M3Z7I5 A0A2M4BRD0 W5J427 A0A2M4CQI9 A0A0T6BD34 A0A182RWS9 A0A0P4VMU7 E0VHX2
A0A194PQX6 A0A0N0PD15 S4P272 A0A2J7RG04 A0A2J7RG25 V5GF00 F4WHG9 A0A2J7RG17 A0A158N933 A0A1Y1KTM5 A0A195FU19 A0A0N0BFX7 A0A067QIZ0 A0A1W4WU45 D6WGH3 E2A6R2 A0A151X4Q2 A0A0J7K897 A0A1J1ISH1 A0A151IQ65 A0A232EV81 K7ITH8 A0A026VY58 A0A088ALI3 E2BSA6 A0A195BUV9 A0A0L7QMJ7 A0A151J863 A0A1I8PMP0 A0A336L1R1 A0A154P2V0 A0A1I8MCX0 A0A1W4VB28 T1PAI8 B4Q3Q1 A0A2A3E8P1 W8BS10 B3MVH7 B4P1Y5 B4IE63 D3TPY9 A0A1A9WKW3 A0A1B0CSC9 B3N3X3 Q9VKB4 A0A1B0FBV4 A0A034WHK6 A0A034WFZ9 A0A1A9UL16 A0A3B0K0U3 A0A0K8VYJ9 A0A0A1WYA7 A0A1A9XCS3 A0A1B0C454 B4N772 A0A1B6KQC3 N6UM87 A0A1L8DG24 Q29NK6 A0A170YNZ4 B4GQK3 A0A1B0D141 A0A069DR55 A0A0L0C6C4 B4KI01 A0A1Q3F1M4 A0A0M4E696 A0A1B6EXZ7 A0A1L8DG36 B4LSF5 B0WPW6 B4JBL6 A0A0A9WF81 A0A224XTC4 A0A1S4F9I7 A0A0V0G8E4 Q17BF6 A0A1B6JQ02 A0A2M4AP89 A0A2M4AKL6 A0A2M4AKI6 A0A182F2A7 A0A2M3Z8G6 A0A2M4BRC4 A0A2M3Z7I5 A0A2M4BRD0 W5J427 A0A2M4CQI9 A0A0T6BD34 A0A182RWS9 A0A0P4VMU7 E0VHX2
PDB
6EM5
E-value=8.29648e-09,
Score=139
Ontologies
Topology
Subcellular location
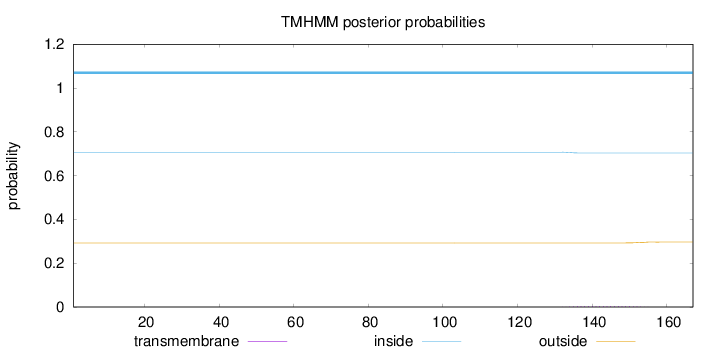
Length:
167
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07294
Exp number, first 60 AAs:
0
Total prob of N-in:
0.70727
inside
1 - 167
Population Genetic Test Statistics
Pi
258.147294
Theta
236.268567
Tajima's D
1.266575
CLR
0.004063
CSRT
0.737263136843158
Interpretation
Uncertain
