Gene
KWMTBOMO08632
Pre Gene Modal
BGIBMGA007630
Annotation
PREDICTED:_methyltransferase-like_protein_6_[Amyelois_transitella]
Full name
Methyltransferase-like protein
Location in the cell
Cytoplasmic Reliability : 2.258
Sequence
CDS
ATGGAAACCGATATTCCTGCGCATGTTGAAGAAACAAGCAGCAGTGACACATTCGTTCACAGGCTGAAAGAACTGTCACAAGAAGAAATAAAGCTCTTACAAAATCAAAATACACGTCTTGTACCCGAAGCCAAAGCGACTAGATTGGAAAAAGATGCTAAACGCCATTGGGATTTATTTTACAAAAGAAACGAAACAAAATTCTTCCGGGACCGGCATTGGACTACACGTGAGTTTCAAGAACTGATTAACTTCGATCCTGAACAGCAAATAGTTTACTTAGAGCTTGGTTGCGGGGTCGGTAATATGATATTCCCGCTGGTCGAAGAAGGCTTTACAAACTTCTTCTTTTACGCGTGTGATTTTTCACCACGTGCAGTAGAATTTGTCAAACGTAACAGCTTGTACGACAAAAACCGCATGAAGGCTTTCTGTGCTGACTTAACCACAGAAGATTTATTCGAAAACATCCAGGAGAATTCAATAGACATCGCGAGTTTGATTTTTGTGCTATCTGCAATCCAGCCGGCTGCTTGGGCCCACGTGGCGGCGCTGGCTTACCGTGCTCTAAAACCCGGGGGCGTATTGCTCTTTAGAGATTATGGGAGGTATGACATGGCCCAGTTACGCTTTAAACCTGGACATAAAATTGCTGACAATTTCTACATGAGACAAGACGGAACTAGGAGCTATTATTTTACGGAAGAAGAGCTACTCAAACTTTTCACTAATGCTGGTTTCATGATCCTCACGAACACGTACGTGCAGCGCAGGACTGAAAATATAAAAAAACCTTTAAATGACTGA
Protein
METDIPAHVEETSSSDTFVHRLKELSQEEIKLLQNQNTRLVPEAKATRLEKDAKRHWDLFYKRNETKFFRDRHWTTREFQELINFDPEQQIVYLELGCGVGNMIFPLVEEGFTNFFFYACDFSPRAVEFVKRNSLYDKNRMKAFCADLTTEDLFENIQENSIDIASLIFVLSAIQPAAWAHVAALAYRALKPGGVLLFRDYGRYDMAQLRFKPGHKIADNFYMRQDGTRSYYFTEEELLKLFTNAGFMILTNTYVQRRTENIKKPLND
Summary
Similarity
Belongs to the methyltransferase superfamily. METL family.
Uniprot
H9JDN3
A0A194PRI7
A0A0L7KQA6
A0A0L7KT11
A0A0N1PGV1
A0A2A4K295
+ More
A0A2H1WXR6 A0A437BXP8 A0A212FB83 A0A0M4ETB7 A0A3B0J1Q2 A0A2M3Z961 A0A0K8TR93 B4KST1 B5E0V3 B4GG35 A0A182H4M8 A0A182F530 B4MDW2 A0A2M4AH06 A0A2M4AGV0 W5J6Y5 A0A182V705 A0A2M4AI12 A0A182TJV1 A0A2M4BV77 T1DL97 A0A084WG42 A0A1B6MLP3 A0A182P318 B0W1C8 A0A1B6L5S3 A0A2M4BVD3 A0A182KTU4 Q16KM2 A0A2M4BV68 A0A453Z0V4 A0A182RHA4 A0A182HRQ6 Q7PK23 A0A1I8M9U9 W8BTU0 A0A1W4VDF5 A0A1I8PUV3 A0A182JDB2 A0A1W4WXH7 A0A0L0CBF4 A0A1B6GJ70 A0A1B6H8H6 B4J3V0 A0A182K991 A0A182LVE8 B4NN56 A0A1A9WI37 A0A0K8VIY4 A0A182WGP8 A0A1A9VPQ7 A0A0L7RAJ5 A0A182NBZ4 D3TRT8 E1ZXQ3 A0A067RUK3 A0A1B0BW20 D6WB49 A0A0P8YC49 A0A0M9A0M6 A0A087ZVL7 A0A182QAC7 A0A154P274 A0A1J1IRR0 A0A336LWN0 A0A1Y1MS57 A0A026WM32 A0A195E1Y7 B3NMM5 A0A2J7QV94 A0A1B6CEB1 K7J9N3 N6UFS3 B4P547 A0A232EFH6 B4HMZ8 B4QBD2 U4URD0 A1ZAY5 A0A0J9RFI9 A0A195CNN9 A0A2M3ZAF5 A0A195FDT4 A0A1A7X1U7 A0A0C9RN84 A0A3Q3B305 U5ERX8 A0A3B4B246 A0A3Q3B2Z7 A0A195B8B0 A0A2A3EQN7 V5IAM4 A0A0E9X9I7 A0A158N9S8
A0A2H1WXR6 A0A437BXP8 A0A212FB83 A0A0M4ETB7 A0A3B0J1Q2 A0A2M3Z961 A0A0K8TR93 B4KST1 B5E0V3 B4GG35 A0A182H4M8 A0A182F530 B4MDW2 A0A2M4AH06 A0A2M4AGV0 W5J6Y5 A0A182V705 A0A2M4AI12 A0A182TJV1 A0A2M4BV77 T1DL97 A0A084WG42 A0A1B6MLP3 A0A182P318 B0W1C8 A0A1B6L5S3 A0A2M4BVD3 A0A182KTU4 Q16KM2 A0A2M4BV68 A0A453Z0V4 A0A182RHA4 A0A182HRQ6 Q7PK23 A0A1I8M9U9 W8BTU0 A0A1W4VDF5 A0A1I8PUV3 A0A182JDB2 A0A1W4WXH7 A0A0L0CBF4 A0A1B6GJ70 A0A1B6H8H6 B4J3V0 A0A182K991 A0A182LVE8 B4NN56 A0A1A9WI37 A0A0K8VIY4 A0A182WGP8 A0A1A9VPQ7 A0A0L7RAJ5 A0A182NBZ4 D3TRT8 E1ZXQ3 A0A067RUK3 A0A1B0BW20 D6WB49 A0A0P8YC49 A0A0M9A0M6 A0A087ZVL7 A0A182QAC7 A0A154P274 A0A1J1IRR0 A0A336LWN0 A0A1Y1MS57 A0A026WM32 A0A195E1Y7 B3NMM5 A0A2J7QV94 A0A1B6CEB1 K7J9N3 N6UFS3 B4P547 A0A232EFH6 B4HMZ8 B4QBD2 U4URD0 A1ZAY5 A0A0J9RFI9 A0A195CNN9 A0A2M3ZAF5 A0A195FDT4 A0A1A7X1U7 A0A0C9RN84 A0A3Q3B305 U5ERX8 A0A3B4B246 A0A3Q3B2Z7 A0A195B8B0 A0A2A3EQN7 V5IAM4 A0A0E9X9I7 A0A158N9S8
EC Number
2.1.1.-
Pubmed
19121390
26354079
26227816
22118469
26369729
17994087
+ More
15632085 26483478 20920257 23761445 24438588 20966253 17510324 12364791 25315136 24495485 26108605 20353571 20798317 24845553 18362917 19820115 28004739 24508170 30249741 20075255 23537049 17550304 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25463417 25613341 21347285
15632085 26483478 20920257 23761445 24438588 20966253 17510324 12364791 25315136 24495485 26108605 20353571 20798317 24845553 18362917 19820115 28004739 24508170 30249741 20075255 23537049 17550304 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25463417 25613341 21347285
EMBL
BABH01003242
KQ459596
KPI95364.1
JTDY01007522
KOB65139.1
JTDY01006222
+ More
KOB66186.1 KQ460497 KPJ14321.1 NWSH01000281 PCG77800.1 ODYU01011836 SOQ57807.1 RSAL01000001 RVE55223.1 AGBW02009363 OWR51004.1 CP012524 ALC40725.1 OUUW01000001 SPP74895.1 GGFM01004312 MBW25063.1 GDAI01000957 JAI16646.1 CH933808 EDW08428.1 CM000071 EDY69425.1 CH479183 EDW35455.1 JXUM01109896 KQ565364 KXJ71047.1 CH940662 EDW58727.2 GGFK01006681 MBW40002.1 GGFK01006690 MBW40011.1 ADMH02002127 ETN58625.1 GGFK01006927 MBW40248.1 GGFJ01007834 MBW56975.1 GAMD01000561 JAB01030.1 ATLV01023437 KE525343 KFB49186.1 GEBQ01003099 JAT36878.1 DS231821 EDS44998.1 GEBQ01020904 JAT19073.1 GGFJ01007823 MBW56964.1 CH477950 EAT34864.1 GGFJ01007824 MBW56965.1 AAAB01008980 APCN01001735 EAA43537.4 GAMC01001785 JAC04771.1 JRES01000655 KNC29562.1 GECZ01007304 JAS62465.1 GECU01036793 JAS70913.1 CH916367 EDW01533.1 AXCM01001182 CH964282 EDW85795.1 GDHF01013465 JAI38849.1 KQ414618 KOC67858.1 CCAG010023811 EZ424140 ADD20416.1 GL435087 EFN74025.1 KK852460 KDR23519.1 JXJN01021578 KQ971311 EEZ98968.1 CH902619 KPU76550.1 KQ435783 KOX74737.1 AXCN02000693 KQ434804 KZC06039.1 CVRI01000057 CRL02410.1 UFQS01000176 UFQT01000176 SSX00779.1 SSX21159.1 GEZM01023287 JAV88523.1 KK107152 QOIP01000007 EZA57097.1 RLU20998.1 KQ979776 KYN19153.1 CH954179 EDV54964.1 NEVH01010478 PNF32503.1 GEDC01025573 JAS11725.1 AAZX01002711 APGK01028666 KB740694 ENN79486.1 CM000158 EDW91748.1 NNAY01005009 OXU17115.1 CH480816 EDW48348.1 CM000362 EDX07556.1 KI207346 ERL95662.1 AE013599 BT030778 AAF57804.3 ABV82160.1 CM002911 KMY94666.1 KQ977532 KYN02092.1 GGFM01004776 MBW25527.1 KQ981656 KYN38546.1 HADW01010394 HADX01000004 SBP11794.1 GBYB01008546 JAG78313.1 GANO01002622 JAB57249.1 KQ976556 KYM80768.1 KZ288193 PBC34103.1 GALX01000930 JAB67536.1 GBXM01010224 JAH98353.1 ADTU01009274
KOB66186.1 KQ460497 KPJ14321.1 NWSH01000281 PCG77800.1 ODYU01011836 SOQ57807.1 RSAL01000001 RVE55223.1 AGBW02009363 OWR51004.1 CP012524 ALC40725.1 OUUW01000001 SPP74895.1 GGFM01004312 MBW25063.1 GDAI01000957 JAI16646.1 CH933808 EDW08428.1 CM000071 EDY69425.1 CH479183 EDW35455.1 JXUM01109896 KQ565364 KXJ71047.1 CH940662 EDW58727.2 GGFK01006681 MBW40002.1 GGFK01006690 MBW40011.1 ADMH02002127 ETN58625.1 GGFK01006927 MBW40248.1 GGFJ01007834 MBW56975.1 GAMD01000561 JAB01030.1 ATLV01023437 KE525343 KFB49186.1 GEBQ01003099 JAT36878.1 DS231821 EDS44998.1 GEBQ01020904 JAT19073.1 GGFJ01007823 MBW56964.1 CH477950 EAT34864.1 GGFJ01007824 MBW56965.1 AAAB01008980 APCN01001735 EAA43537.4 GAMC01001785 JAC04771.1 JRES01000655 KNC29562.1 GECZ01007304 JAS62465.1 GECU01036793 JAS70913.1 CH916367 EDW01533.1 AXCM01001182 CH964282 EDW85795.1 GDHF01013465 JAI38849.1 KQ414618 KOC67858.1 CCAG010023811 EZ424140 ADD20416.1 GL435087 EFN74025.1 KK852460 KDR23519.1 JXJN01021578 KQ971311 EEZ98968.1 CH902619 KPU76550.1 KQ435783 KOX74737.1 AXCN02000693 KQ434804 KZC06039.1 CVRI01000057 CRL02410.1 UFQS01000176 UFQT01000176 SSX00779.1 SSX21159.1 GEZM01023287 JAV88523.1 KK107152 QOIP01000007 EZA57097.1 RLU20998.1 KQ979776 KYN19153.1 CH954179 EDV54964.1 NEVH01010478 PNF32503.1 GEDC01025573 JAS11725.1 AAZX01002711 APGK01028666 KB740694 ENN79486.1 CM000158 EDW91748.1 NNAY01005009 OXU17115.1 CH480816 EDW48348.1 CM000362 EDX07556.1 KI207346 ERL95662.1 AE013599 BT030778 AAF57804.3 ABV82160.1 CM002911 KMY94666.1 KQ977532 KYN02092.1 GGFM01004776 MBW25527.1 KQ981656 KYN38546.1 HADW01010394 HADX01000004 SBP11794.1 GBYB01008546 JAG78313.1 GANO01002622 JAB57249.1 KQ976556 KYM80768.1 KZ288193 PBC34103.1 GALX01000930 JAB67536.1 GBXM01010224 JAH98353.1 ADTU01009274
Proteomes
UP000005204
UP000053268
UP000037510
UP000053240
UP000218220
UP000283053
+ More
UP000007151 UP000092553 UP000268350 UP000009192 UP000001819 UP000008744 UP000069940 UP000249989 UP000069272 UP000008792 UP000000673 UP000075903 UP000075902 UP000030765 UP000075885 UP000002320 UP000075882 UP000008820 UP000075900 UP000075840 UP000007062 UP000095301 UP000192221 UP000095300 UP000075880 UP000192223 UP000037069 UP000001070 UP000075881 UP000075883 UP000007798 UP000091820 UP000075920 UP000078200 UP000053825 UP000075884 UP000092444 UP000000311 UP000027135 UP000092460 UP000007266 UP000007801 UP000053105 UP000005203 UP000075886 UP000076502 UP000183832 UP000053097 UP000279307 UP000078492 UP000008711 UP000235965 UP000002358 UP000019118 UP000002282 UP000215335 UP000001292 UP000000304 UP000030742 UP000000803 UP000078542 UP000078541 UP000264800 UP000261520 UP000078540 UP000242457 UP000005205
UP000007151 UP000092553 UP000268350 UP000009192 UP000001819 UP000008744 UP000069940 UP000249989 UP000069272 UP000008792 UP000000673 UP000075903 UP000075902 UP000030765 UP000075885 UP000002320 UP000075882 UP000008820 UP000075900 UP000075840 UP000007062 UP000095301 UP000192221 UP000095300 UP000075880 UP000192223 UP000037069 UP000001070 UP000075881 UP000075883 UP000007798 UP000091820 UP000075920 UP000078200 UP000053825 UP000075884 UP000092444 UP000000311 UP000027135 UP000092460 UP000007266 UP000007801 UP000053105 UP000005203 UP000075886 UP000076502 UP000183832 UP000053097 UP000279307 UP000078492 UP000008711 UP000235965 UP000002358 UP000019118 UP000002282 UP000215335 UP000001292 UP000000304 UP000030742 UP000000803 UP000078542 UP000078541 UP000264800 UP000261520 UP000078540 UP000242457 UP000005205
Pfam
Interpro
SUPFAM
SSF53335
SSF53335
Gene 3D
CDD
ProteinModelPortal
H9JDN3
A0A194PRI7
A0A0L7KQA6
A0A0L7KT11
A0A0N1PGV1
A0A2A4K295
+ More
A0A2H1WXR6 A0A437BXP8 A0A212FB83 A0A0M4ETB7 A0A3B0J1Q2 A0A2M3Z961 A0A0K8TR93 B4KST1 B5E0V3 B4GG35 A0A182H4M8 A0A182F530 B4MDW2 A0A2M4AH06 A0A2M4AGV0 W5J6Y5 A0A182V705 A0A2M4AI12 A0A182TJV1 A0A2M4BV77 T1DL97 A0A084WG42 A0A1B6MLP3 A0A182P318 B0W1C8 A0A1B6L5S3 A0A2M4BVD3 A0A182KTU4 Q16KM2 A0A2M4BV68 A0A453Z0V4 A0A182RHA4 A0A182HRQ6 Q7PK23 A0A1I8M9U9 W8BTU0 A0A1W4VDF5 A0A1I8PUV3 A0A182JDB2 A0A1W4WXH7 A0A0L0CBF4 A0A1B6GJ70 A0A1B6H8H6 B4J3V0 A0A182K991 A0A182LVE8 B4NN56 A0A1A9WI37 A0A0K8VIY4 A0A182WGP8 A0A1A9VPQ7 A0A0L7RAJ5 A0A182NBZ4 D3TRT8 E1ZXQ3 A0A067RUK3 A0A1B0BW20 D6WB49 A0A0P8YC49 A0A0M9A0M6 A0A087ZVL7 A0A182QAC7 A0A154P274 A0A1J1IRR0 A0A336LWN0 A0A1Y1MS57 A0A026WM32 A0A195E1Y7 B3NMM5 A0A2J7QV94 A0A1B6CEB1 K7J9N3 N6UFS3 B4P547 A0A232EFH6 B4HMZ8 B4QBD2 U4URD0 A1ZAY5 A0A0J9RFI9 A0A195CNN9 A0A2M3ZAF5 A0A195FDT4 A0A1A7X1U7 A0A0C9RN84 A0A3Q3B305 U5ERX8 A0A3B4B246 A0A3Q3B2Z7 A0A195B8B0 A0A2A3EQN7 V5IAM4 A0A0E9X9I7 A0A158N9S8
A0A2H1WXR6 A0A437BXP8 A0A212FB83 A0A0M4ETB7 A0A3B0J1Q2 A0A2M3Z961 A0A0K8TR93 B4KST1 B5E0V3 B4GG35 A0A182H4M8 A0A182F530 B4MDW2 A0A2M4AH06 A0A2M4AGV0 W5J6Y5 A0A182V705 A0A2M4AI12 A0A182TJV1 A0A2M4BV77 T1DL97 A0A084WG42 A0A1B6MLP3 A0A182P318 B0W1C8 A0A1B6L5S3 A0A2M4BVD3 A0A182KTU4 Q16KM2 A0A2M4BV68 A0A453Z0V4 A0A182RHA4 A0A182HRQ6 Q7PK23 A0A1I8M9U9 W8BTU0 A0A1W4VDF5 A0A1I8PUV3 A0A182JDB2 A0A1W4WXH7 A0A0L0CBF4 A0A1B6GJ70 A0A1B6H8H6 B4J3V0 A0A182K991 A0A182LVE8 B4NN56 A0A1A9WI37 A0A0K8VIY4 A0A182WGP8 A0A1A9VPQ7 A0A0L7RAJ5 A0A182NBZ4 D3TRT8 E1ZXQ3 A0A067RUK3 A0A1B0BW20 D6WB49 A0A0P8YC49 A0A0M9A0M6 A0A087ZVL7 A0A182QAC7 A0A154P274 A0A1J1IRR0 A0A336LWN0 A0A1Y1MS57 A0A026WM32 A0A195E1Y7 B3NMM5 A0A2J7QV94 A0A1B6CEB1 K7J9N3 N6UFS3 B4P547 A0A232EFH6 B4HMZ8 B4QBD2 U4URD0 A1ZAY5 A0A0J9RFI9 A0A195CNN9 A0A2M3ZAF5 A0A195FDT4 A0A1A7X1U7 A0A0C9RN84 A0A3Q3B305 U5ERX8 A0A3B4B246 A0A3Q3B2Z7 A0A195B8B0 A0A2A3EQN7 V5IAM4 A0A0E9X9I7 A0A158N9S8
PDB
5WP4
E-value=0.00108479,
Score=98
Ontologies
PANTHER
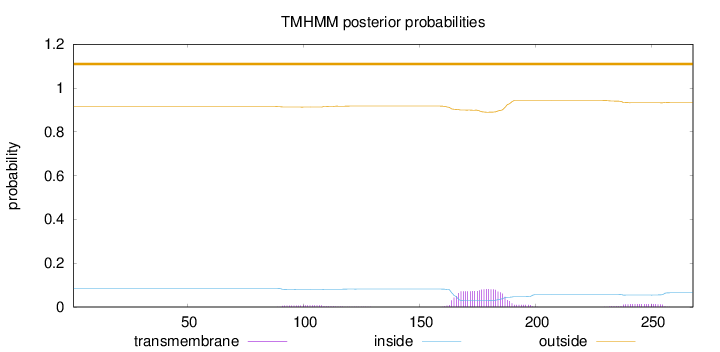
Topology
Length:
268
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.22117
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08502
outside
1 - 268
Population Genetic Test Statistics
Pi
280.462767
Theta
214.188935
Tajima's D
0.980019
CLR
0
CSRT
0.654917254137293
Interpretation
Uncertain
