Gene
KWMTBOMO08629 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007956
Annotation
PREDICTED:_innexin_inx7_[Papilio_xuthus]
Full name
Innexin
+ More
Innexin inx7
Innexin inx7
Alternative Name
Gap junction protein prp7
Pas-related protein 7
Pas-related protein 7
Location in the cell
PlasmaMembrane Reliability : 4.516
Sequence
CDS
ATGTTAGTCGCGTCGCTTAACAGTCTCTCGCCGCGGCTGCGGTTCCAATTTAGCAAACCGAAGATCGAGAATGTGGCCTTCAGGCTGCATTACCAGCTGACTGTGACGATTTTGCTGGCTTTCGTGATCCTGGTCTGCGCTAGAGAGTATTTCGGGGATCACATCAAATGTCTTTCGGATCAGGGTGTGCCCGACCATGTAATTCAGACGTACTGCTTCTTCATGGCCACTTTTACAATAGTTCGTCATTACAACGAGAGTCTACTTCAAGGAGAGTTCCTGCCTCATCCCGGAGTAGGGCCCATCCTTGCCACAGACGAGACCATCCACCACACGTATTACCAGTGGGTACCTTTCGTACTCTTCATCCAGTCTATATGCTTCTACCTGCCTCATTACATATGGAAGACAAAAGAAGGTGGCAGGATAAAGGCCCTGGTTGACGGACTCCAGTACGCCGGACTGGCGCTCCACGACGACGACATCACCGTCAACGGCACCACAGTACCTTCAAAGAAGACCCTCGAAAACAAACTGGATTCAATTAGGAAAGACATCATATTAAGGTTGAAGATATCGAGGACGTGGTCGACTTGGCTCGTCTCCATGGAGGTCGCGAATCTGCTGCACGTCATGTTCCAGGTGTGGGTGATCAACAAGTTCCTAAACGGGGCGTTCATGTCACTAGGCCCCAGAGTACTCGAAACGAAAGACTGGAGCCACATAGTCGATCCGCTGGAGCTTGTGTTTCCGAAGGTAACGAAGTGCATCTTCCACAAGTACGGTCCGAGTGGCTCAATCCAGCAACACGACGCGCTCTGCGTAATGGCCCTCAATATAATCCACGAGAAGATCTACACAGTGCTTTGGTTCTGGCTGCTTTTCCTTTTCATTGTCTCCTTATTGGCTGTGATATGGCGGGTAATTTCGTTTTTCCTGTACCGCCGGTCGCTGCGGTTCAACGAGATGATGTTCCGTCACGTGAGCAACGCCAAGTTCAACCCGTACAACGTCATCAGGGTGGTCAACGGCTGCGAGTTCGCCGATTGGCTCTTCCTCTATTACCTTGCGAAGAATATGCAAGGCTTTGTATTCCAGGCACTATTTGTGCGATTAGCGGAGGAGTTGGAGAAAAGAGAACTGCCTTACGATGACCAAGGCACGGAGGAGAAGGGAGCTGAACCAATATTAGTCGGAAAAGTAGATATTGACGACGAAACTATGCCGTTAAAAAGAGATAAGAAATCATCGTGA
Protein
MLVASLNSLSPRLRFQFSKPKIENVAFRLHYQLTVTILLAFVILVCAREYFGDHIKCLSDQGVPDHVIQTYCFFMATFTIVRHYNESLLQGEFLPHPGVGPILATDETIHHTYYQWVPFVLFIQSICFYLPHYIWKTKEGGRIKALVDGLQYAGLALHDDDITVNGTTVPSKKTLENKLDSIRKDIILRLKISRTWSTWLVSMEVANLLHVMFQVWVINKFLNGAFMSLGPRVLETKDWSHIVDPLELVFPKVTKCIFHKYGPSGSIQQHDALCVMALNIIHEKIYTVLWFWLLFLFIVSLLAVIWRVISFFLYRRSLRFNEMMFRHVSNAKFNPYNVIRVVNGCEFADWLFLYYLAKNMQGFVFQALFVRLAEELEKRELPYDDQGTEEKGAEPILVGKVDIDDETMPLKRDKKSS
Summary
Description
Structural component of the gap junctions.
Structural components of the gap junctions.
Structural components of the gap junctions.
Similarity
Belongs to the pannexin family.
Keywords
Alternative splicing
Cell junction
Cell membrane
Complete proteome
Gap junction
Ion channel
Ion transport
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Innexin
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A1E1W3D1
A0A0N1I9S5
A0A194PW50
A0A212FCE9
A0A3S2NSG4
A0A2A4K2D6
+ More
A0A3G1PLL7 A0A2H1WV22 A0A3S2P8V0 A0A232ELL2 E1ZXF0 A0A1Y1L4F3 A0A195D3Q4 A0A158NQK5 U4UG91 A0A195BR20 N6T152 A0A026WV24 A0A151XB47 A0A195EQV2 F4WK99 K7J6Q9 E2BMI4 A0A1Y1KBU2 A0A3L8E3E4 A0A0M9A1U3 A0A154NXH2 A0A088A2D0 A0A0L7R3G8 A0A0L0BUC5 A0A0C9R0I7 A0A2J7PHB7 A0A182QUM8 A0A067QS42 A0A0A1XPB7 A0A1I8M693 A0A1A9WPJ8 A0A0K8UU33 A0A182VWC8 T1PCB4 A0A182R518 A0A1L8EHW0 A0A0C9QE81 A0A182U464 D3TPD3 A0A182XA19 A0A1B0G4W3 A0A182L9B3 A0A182HKZ8 Q7PKP9 A0A1A9UUN4 A0A182N9Z5 A0A1A9ZE60 W8BD06 A0A182V3Z4 A0A182MQQ6 A0A034VI85 A0A1I8P5M9 A0A182PH09 A0A195DI22 A0A182SBJ0 A0A0C9R8L9 A0A1Q3FKV1 Q16YE3 A0A182YBA4 W5J902 A0A182FRS8 A0A1B0DD11 A0A1L8E339 A0A336MRI1 A0A336M5N8 A0A336LYL1 A0A1B0CW53 A0A084VW21 A0A182JAI9 A0A0T6BB60 A0A0L7KPR2 A0A0C9R3X1 A0A1B0AP74 A0A1A9XRV5 A0A2A3E9W3 J9K6I2 A0A1J1HXG5 B4Q1C7 B4L564 D6X4I9 B3MQ10 B4IHI5 B4R5W3 A0A139WAT1 B0WZV4 Q29GY9 Q9V3W6 B4M9W3 A0A3B0K809 B4JMT0 B4NEF9 B4H2D3
A0A3G1PLL7 A0A2H1WV22 A0A3S2P8V0 A0A232ELL2 E1ZXF0 A0A1Y1L4F3 A0A195D3Q4 A0A158NQK5 U4UG91 A0A195BR20 N6T152 A0A026WV24 A0A151XB47 A0A195EQV2 F4WK99 K7J6Q9 E2BMI4 A0A1Y1KBU2 A0A3L8E3E4 A0A0M9A1U3 A0A154NXH2 A0A088A2D0 A0A0L7R3G8 A0A0L0BUC5 A0A0C9R0I7 A0A2J7PHB7 A0A182QUM8 A0A067QS42 A0A0A1XPB7 A0A1I8M693 A0A1A9WPJ8 A0A0K8UU33 A0A182VWC8 T1PCB4 A0A182R518 A0A1L8EHW0 A0A0C9QE81 A0A182U464 D3TPD3 A0A182XA19 A0A1B0G4W3 A0A182L9B3 A0A182HKZ8 Q7PKP9 A0A1A9UUN4 A0A182N9Z5 A0A1A9ZE60 W8BD06 A0A182V3Z4 A0A182MQQ6 A0A034VI85 A0A1I8P5M9 A0A182PH09 A0A195DI22 A0A182SBJ0 A0A0C9R8L9 A0A1Q3FKV1 Q16YE3 A0A182YBA4 W5J902 A0A182FRS8 A0A1B0DD11 A0A1L8E339 A0A336MRI1 A0A336M5N8 A0A336LYL1 A0A1B0CW53 A0A084VW21 A0A182JAI9 A0A0T6BB60 A0A0L7KPR2 A0A0C9R3X1 A0A1B0AP74 A0A1A9XRV5 A0A2A3E9W3 J9K6I2 A0A1J1HXG5 B4Q1C7 B4L564 D6X4I9 B3MQ10 B4IHI5 B4R5W3 A0A139WAT1 B0WZV4 Q29GY9 Q9V3W6 B4M9W3 A0A3B0K809 B4JMT0 B4NEF9 B4H2D3
Pubmed
26354079
22118469
29361596
28648823
20798317
28004739
+ More
21347285 23537049 24508170 21719571 20075255 30249741 26108605 24845553 25830018 25315136 20353571 20966253 12364791 14747013 17210077 24495485 25348373 17510324 25585357 25244985 20920257 23761445 24438588 26227816 17994087 17550304 18362917 19820115 15632085 10352230 10731132 12537572 12537569
21347285 23537049 24508170 21719571 20075255 30249741 26108605 24845553 25830018 25315136 20353571 20966253 12364791 14747013 17210077 24495485 25348373 17510324 25585357 25244985 20920257 23761445 24438588 26227816 17994087 17550304 18362917 19820115 15632085 10352230 10731132 12537572 12537569
EMBL
GDQN01009566
GDQN01005453
GDQN01001937
JAT81488.1
JAT85601.1
JAT89117.1
+ More
KQ460497 KPJ14318.1 KQ459596 KPI95360.1 AGBW02009200 OWR51393.1 RSAL01000001 RVE55225.1 NWSH01000281 PCG77802.1 MG000945 AVM81263.1 ODYU01010875 SOQ56254.1 RVE55226.1 NNAY01003541 OXU19235.1 GL435038 EFN74127.1 GEZM01065084 JAV68563.1 KQ976885 KYN07540.1 ADTU01023369 KB632184 ERL89626.1 KQ976419 KYM89301.1 APGK01048500 KB741112 ENN73824.1 KK107087 EZA59867.1 KQ982335 KYQ57518.1 KQ982021 KYN30556.1 GL888199 EGI65323.1 AAZX01003107 GL449233 EFN83093.1 GEZM01091116 GEZM01091114 JAV57085.1 QOIP01000001 RLU26915.1 KQ435762 KOX75477.1 KQ434769 KZC03794.1 KQ414663 KOC65364.1 JRES01001325 KNC23626.1 GBYB01001505 JAG71272.1 NEVH01025142 PNF15719.1 AXCN02001521 KK853092 KDR11526.1 GBXI01001461 JAD12831.1 GDHF01022115 JAI30199.1 KA646329 AFP60958.1 GFDG01000593 JAV18206.1 GBYB01012713 GBYB01012716 JAG82480.1 JAG82483.1 EZ423285 ADD19561.1 CCAG010000781 APCN01000876 AAAB01008987 EAA43180.3 GAMC01009999 GAMC01009998 JAB96557.1 AXCM01008205 GAKP01015931 JAC43021.1 KQ980824 KYN12491.1 GBYB01012714 JAG82481.1 GFDL01006838 JAV28207.1 KJ736827 CH477518 AIF75099.1 EAT39624.1 ADMH02001846 ETN60917.1 AJVK01014292 AJVK01014293 GFDF01000947 JAV13137.1 UFQT01002269 SSX33052.1 UFQT01000188 SSX21348.1 SSX21347.1 AJWK01031778 ATLV01017403 KE525167 KFB42165.1 LJIG01002374 KRT84566.1 JTDY01007762 KOB64959.1 GBYB01001503 GBYB01001504 GBYB01001506 JAG71270.1 JAG71271.1 JAG71273.1 JXJN01001234 KZ288311 PBC28517.1 ABLF02020775 CVRI01000024 CRK92218.1 CM000162 EDX02416.1 CH933811 EDW06323.1 KQ971380 EEZ97261.2 CH902621 EDV44436.1 CH480839 EDW49336.1 CM000366 EDX17299.1 KYB25015.1 DS232218 EDS37779.1 CH379064 EAL31970.1 AF137270 AE014298 AY051561 BT099510 CH940655 EDW66022.1 OUUW01000016 SPP88802.1 CH916371 EDV92023.1 CH964239 EDW82128.1 CH479204 EDW30500.1
KQ460497 KPJ14318.1 KQ459596 KPI95360.1 AGBW02009200 OWR51393.1 RSAL01000001 RVE55225.1 NWSH01000281 PCG77802.1 MG000945 AVM81263.1 ODYU01010875 SOQ56254.1 RVE55226.1 NNAY01003541 OXU19235.1 GL435038 EFN74127.1 GEZM01065084 JAV68563.1 KQ976885 KYN07540.1 ADTU01023369 KB632184 ERL89626.1 KQ976419 KYM89301.1 APGK01048500 KB741112 ENN73824.1 KK107087 EZA59867.1 KQ982335 KYQ57518.1 KQ982021 KYN30556.1 GL888199 EGI65323.1 AAZX01003107 GL449233 EFN83093.1 GEZM01091116 GEZM01091114 JAV57085.1 QOIP01000001 RLU26915.1 KQ435762 KOX75477.1 KQ434769 KZC03794.1 KQ414663 KOC65364.1 JRES01001325 KNC23626.1 GBYB01001505 JAG71272.1 NEVH01025142 PNF15719.1 AXCN02001521 KK853092 KDR11526.1 GBXI01001461 JAD12831.1 GDHF01022115 JAI30199.1 KA646329 AFP60958.1 GFDG01000593 JAV18206.1 GBYB01012713 GBYB01012716 JAG82480.1 JAG82483.1 EZ423285 ADD19561.1 CCAG010000781 APCN01000876 AAAB01008987 EAA43180.3 GAMC01009999 GAMC01009998 JAB96557.1 AXCM01008205 GAKP01015931 JAC43021.1 KQ980824 KYN12491.1 GBYB01012714 JAG82481.1 GFDL01006838 JAV28207.1 KJ736827 CH477518 AIF75099.1 EAT39624.1 ADMH02001846 ETN60917.1 AJVK01014292 AJVK01014293 GFDF01000947 JAV13137.1 UFQT01002269 SSX33052.1 UFQT01000188 SSX21348.1 SSX21347.1 AJWK01031778 ATLV01017403 KE525167 KFB42165.1 LJIG01002374 KRT84566.1 JTDY01007762 KOB64959.1 GBYB01001503 GBYB01001504 GBYB01001506 JAG71270.1 JAG71271.1 JAG71273.1 JXJN01001234 KZ288311 PBC28517.1 ABLF02020775 CVRI01000024 CRK92218.1 CM000162 EDX02416.1 CH933811 EDW06323.1 KQ971380 EEZ97261.2 CH902621 EDV44436.1 CH480839 EDW49336.1 CM000366 EDX17299.1 KYB25015.1 DS232218 EDS37779.1 CH379064 EAL31970.1 AF137270 AE014298 AY051561 BT099510 CH940655 EDW66022.1 OUUW01000016 SPP88802.1 CH916371 EDV92023.1 CH964239 EDW82128.1 CH479204 EDW30500.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000283053
UP000218220
UP000215335
+ More
UP000000311 UP000078542 UP000005205 UP000030742 UP000078540 UP000019118 UP000053097 UP000075809 UP000078541 UP000007755 UP000002358 UP000008237 UP000279307 UP000053105 UP000076502 UP000005203 UP000053825 UP000037069 UP000235965 UP000075886 UP000027135 UP000095301 UP000091820 UP000075920 UP000075900 UP000075902 UP000076407 UP000092444 UP000075882 UP000075840 UP000007062 UP000078200 UP000075884 UP000092445 UP000075903 UP000075883 UP000095300 UP000075885 UP000078492 UP000075901 UP000008820 UP000076408 UP000000673 UP000069272 UP000092462 UP000092461 UP000030765 UP000075880 UP000037510 UP000092460 UP000092443 UP000242457 UP000007819 UP000183832 UP000002282 UP000009192 UP000007266 UP000007801 UP000001292 UP000000304 UP000002320 UP000001819 UP000000803 UP000008792 UP000268350 UP000001070 UP000007798 UP000008744
UP000000311 UP000078542 UP000005205 UP000030742 UP000078540 UP000019118 UP000053097 UP000075809 UP000078541 UP000007755 UP000002358 UP000008237 UP000279307 UP000053105 UP000076502 UP000005203 UP000053825 UP000037069 UP000235965 UP000075886 UP000027135 UP000095301 UP000091820 UP000075920 UP000075900 UP000075902 UP000076407 UP000092444 UP000075882 UP000075840 UP000007062 UP000078200 UP000075884 UP000092445 UP000075903 UP000075883 UP000095300 UP000075885 UP000078492 UP000075901 UP000008820 UP000076408 UP000000673 UP000069272 UP000092462 UP000092461 UP000030765 UP000075880 UP000037510 UP000092460 UP000092443 UP000242457 UP000007819 UP000183832 UP000002282 UP000009192 UP000007266 UP000007801 UP000001292 UP000000304 UP000002320 UP000001819 UP000000803 UP000008792 UP000268350 UP000001070 UP000007798 UP000008744
PRIDE
Pfam
PF00876 Innexin
Interpro
IPR000990
Innexin
ProteinModelPortal
A0A1E1W3D1
A0A0N1I9S5
A0A194PW50
A0A212FCE9
A0A3S2NSG4
A0A2A4K2D6
+ More
A0A3G1PLL7 A0A2H1WV22 A0A3S2P8V0 A0A232ELL2 E1ZXF0 A0A1Y1L4F3 A0A195D3Q4 A0A158NQK5 U4UG91 A0A195BR20 N6T152 A0A026WV24 A0A151XB47 A0A195EQV2 F4WK99 K7J6Q9 E2BMI4 A0A1Y1KBU2 A0A3L8E3E4 A0A0M9A1U3 A0A154NXH2 A0A088A2D0 A0A0L7R3G8 A0A0L0BUC5 A0A0C9R0I7 A0A2J7PHB7 A0A182QUM8 A0A067QS42 A0A0A1XPB7 A0A1I8M693 A0A1A9WPJ8 A0A0K8UU33 A0A182VWC8 T1PCB4 A0A182R518 A0A1L8EHW0 A0A0C9QE81 A0A182U464 D3TPD3 A0A182XA19 A0A1B0G4W3 A0A182L9B3 A0A182HKZ8 Q7PKP9 A0A1A9UUN4 A0A182N9Z5 A0A1A9ZE60 W8BD06 A0A182V3Z4 A0A182MQQ6 A0A034VI85 A0A1I8P5M9 A0A182PH09 A0A195DI22 A0A182SBJ0 A0A0C9R8L9 A0A1Q3FKV1 Q16YE3 A0A182YBA4 W5J902 A0A182FRS8 A0A1B0DD11 A0A1L8E339 A0A336MRI1 A0A336M5N8 A0A336LYL1 A0A1B0CW53 A0A084VW21 A0A182JAI9 A0A0T6BB60 A0A0L7KPR2 A0A0C9R3X1 A0A1B0AP74 A0A1A9XRV5 A0A2A3E9W3 J9K6I2 A0A1J1HXG5 B4Q1C7 B4L564 D6X4I9 B3MQ10 B4IHI5 B4R5W3 A0A139WAT1 B0WZV4 Q29GY9 Q9V3W6 B4M9W3 A0A3B0K809 B4JMT0 B4NEF9 B4H2D3
A0A3G1PLL7 A0A2H1WV22 A0A3S2P8V0 A0A232ELL2 E1ZXF0 A0A1Y1L4F3 A0A195D3Q4 A0A158NQK5 U4UG91 A0A195BR20 N6T152 A0A026WV24 A0A151XB47 A0A195EQV2 F4WK99 K7J6Q9 E2BMI4 A0A1Y1KBU2 A0A3L8E3E4 A0A0M9A1U3 A0A154NXH2 A0A088A2D0 A0A0L7R3G8 A0A0L0BUC5 A0A0C9R0I7 A0A2J7PHB7 A0A182QUM8 A0A067QS42 A0A0A1XPB7 A0A1I8M693 A0A1A9WPJ8 A0A0K8UU33 A0A182VWC8 T1PCB4 A0A182R518 A0A1L8EHW0 A0A0C9QE81 A0A182U464 D3TPD3 A0A182XA19 A0A1B0G4W3 A0A182L9B3 A0A182HKZ8 Q7PKP9 A0A1A9UUN4 A0A182N9Z5 A0A1A9ZE60 W8BD06 A0A182V3Z4 A0A182MQQ6 A0A034VI85 A0A1I8P5M9 A0A182PH09 A0A195DI22 A0A182SBJ0 A0A0C9R8L9 A0A1Q3FKV1 Q16YE3 A0A182YBA4 W5J902 A0A182FRS8 A0A1B0DD11 A0A1L8E339 A0A336MRI1 A0A336M5N8 A0A336LYL1 A0A1B0CW53 A0A084VW21 A0A182JAI9 A0A0T6BB60 A0A0L7KPR2 A0A0C9R3X1 A0A1B0AP74 A0A1A9XRV5 A0A2A3E9W3 J9K6I2 A0A1J1HXG5 B4Q1C7 B4L564 D6X4I9 B3MQ10 B4IHI5 B4R5W3 A0A139WAT1 B0WZV4 Q29GY9 Q9V3W6 B4M9W3 A0A3B0K809 B4JMT0 B4NEF9 B4H2D3
PDB
5H1R
E-value=5.83309e-15,
Score=197
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
Cell junction
Gap junction
Cell junction
Gap junction
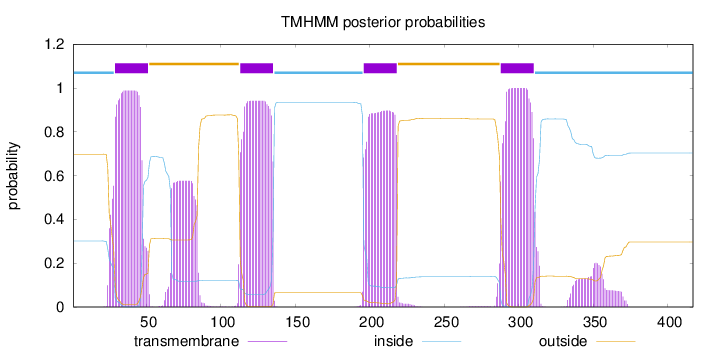
Length:
417
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
100.36733
Exp number, first 60 AAs:
21.19256
Total prob of N-in:
0.30194
POSSIBLE N-term signal
sequence
inside
1 - 28
TMhelix
29 - 51
outside
52 - 112
TMhelix
113 - 135
inside
136 - 195
TMhelix
196 - 218
outside
219 - 287
TMhelix
288 - 310
inside
311 - 417
Population Genetic Test Statistics
Pi
207.505214
Theta
180.873097
Tajima's D
2.058028
CLR
0.641524
CSRT
0.890555472226389
Interpretation
Uncertain
