Gene
KWMTBOMO08627 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007628
Annotation
PREDICTED:_uncharacterized_protein_LOC106708416_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 3.613
Sequence
CDS
ATGTCGTTGAAAACGCTAGTCTTCTGTGTTTGCGTTACTTTAGCTTCGGCGGCGATACAGCGCTCTGCTTTGGAGCCAATCCCTGATAAATTTAGTGGCCTTCAAGGCTGTTACATCAGGGAGTTCGATGGTGTCCTTCCCTTTGATTCTGTAACATACTCGAAGGAAGCTGAATGCACCCGATACTTCTGCGCGCACGATGCTGTCATACACCAATCGTGTAAAGATGAGCTGAGCACAGACACCCCTGTATGTACCTTTACCAAAACAGAAGAAAAACGTGACACAATCTGCATTCTTTGA
Protein
MSLKTLVFCVCVTLASAAIQRSALEPIPDKFSGLQGCYIREFDGVLPFDSVTYSKEAECTRYFCAHDAVIHQSCKDELSTDTPVCTFTKTEEKRDTICIL
Summary
Uniprot
EMBL
Proteomes
Pfam
PF15430 SVWC
Interpro
IPR029277
SVWC_dom
ProteinModelPortal
Ontologies
GO
Topology
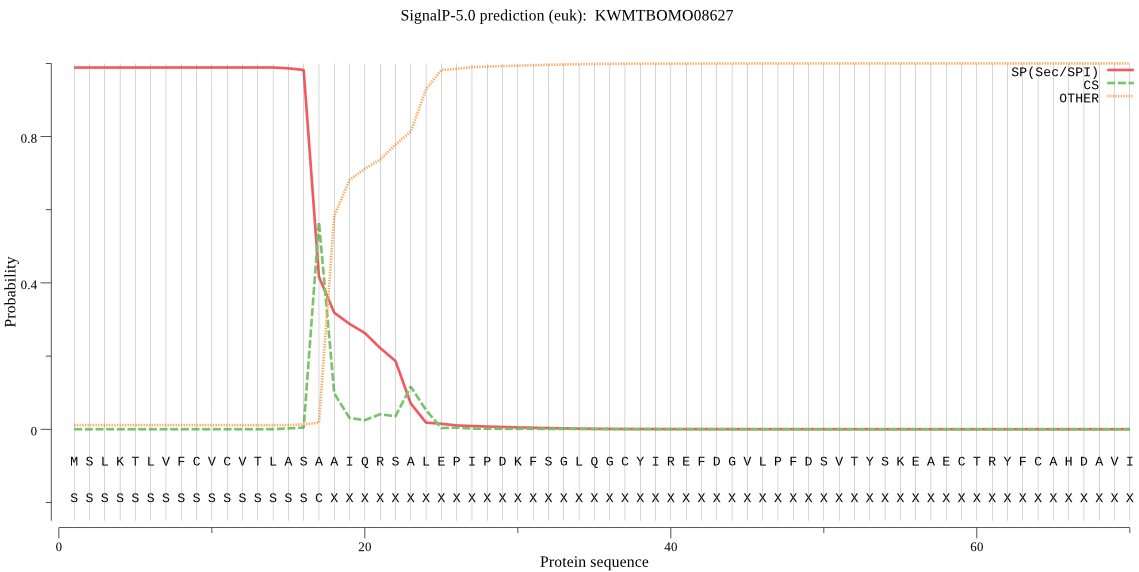
SignalP
Position: 1 - 17,
Likelihood: 0.988474
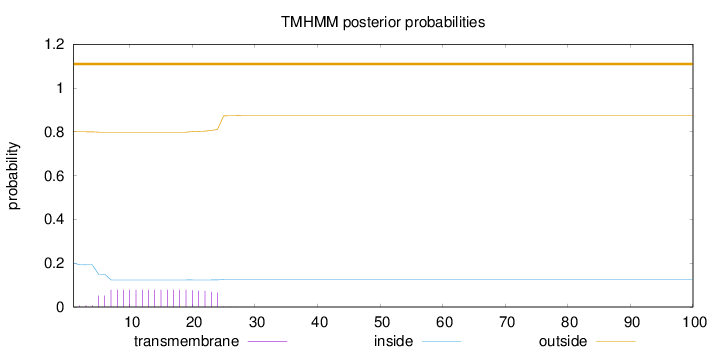
Length:
100
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.50189
Exp number, first 60 AAs:
1.50189
Total prob of N-in:
0.19994
outside
1 - 100
Population Genetic Test Statistics
Pi
241.595002
Theta
159.867251
Tajima's D
1.803605
CLR
0.987519
CSRT
0.849557522123894
Interpretation
Uncertain
