Gene
KWMTBOMO08624
Pre Gene Modal
BGIBMGA007626
Annotation
PREDICTED:_innexin_shaking-B_isoform_X1_[Bombyx_mori]
Full name
Innexin
+ More
Innexin shaking-B
Innexin shaking-B
Alternative Name
Protein passover
Location in the cell
PlasmaMembrane Reliability : 3.988
Sequence
CDS
ATGTTAGACGTATTCCGCGGTTTGAAGAATCTAATTAAGGTGAACTATGTGCATATCGACTCGCCTGTATTCCGGTTGCATTACTCGATCACGGTCATCCTGCTGATATCGTTCAGTCTCATTGTGACCACGCGCCAGTACGTCGGGAACCCCATAGACTGTATCCACACCAAGGACATCCCAGAGGACGTTCTGAACACGTACTGCTGGATACATTCGACATACACCCTCAAGAGCTACTTCGAGAAGACGGTGGGCGTCGAGGTGCCGTACCCTGGGATAGGGAACAGCAGGGAGAAGGGCAAGGAAGATATGAGCGACAGGAAAGTCTACAAGTACTACCAATGGGTCTGCTTCTGTCTTTTTTTTCAGGCAATGCTGTTCTACGCCCCGCGGTGGCTGTGGAAATCTTGGGAGGGCGGCAAGATCCGCGCCCTCATGATGGACCTAGACGTCGCAGTCTGCACGGAGATAGAGAAGAAGACAAAAAAGAAACTAATCCTGGACTATCTGTGGGAAAACCTGAGGTACCACAACTGGTGGGCCTACAGGTACTACCTGTGCGAAGCGCTTGCCCTTGTCAATGTTATAGGGCAAATGTTCTTGATGAACAGGTTCTTCGACGGAGAGTTTATGACCTTCGGGCTGAAGGTGATCAAGTACATGGAGTCCGACCAGGAGATACGAATCGACCCCATGATATACATATTTCCAAGGATGGTGAAGTGTACGCTTTTCAACAAATTCGGTTCATCCGGCGAGGTGGAGCGGCACGACGCGCTCTGCATCTTGCCCCTGAACGTAGTCAACGAGAAGATCTACATCTTCCTCTGGTTCTGGTTCGTCATCCTCGGCTTCCTTACTTTCATCACGCTCATCTACAGGATCATCATCATCTTCTCGCCGCGCATGCGCGTCTACATGATGCGAATGAGGTTCCGACTCGTGAGGCGGGACAATATCGACACCATAGTGAGGAGAAGCAAGATGGGAGACTGGTATCTGCTCTACATACTAGGAGAGAACCTCGACTCGATCATCTTCAGGGATATAATGCAGGAGTTCGCGAACAAGCTGAACCACAACTACCAGCACCACATTCACGGCGTCCCGGACGCGTGA
Protein
MLDVFRGLKNLIKVNYVHIDSPVFRLHYSITVILLISFSLIVTTRQYVGNPIDCIHTKDIPEDVLNTYCWIHSTYTLKSYFEKTVGVEVPYPGIGNSREKGKEDMSDRKVYKYYQWVCFCLFFQAMLFYAPRWLWKSWEGGKIRALMMDLDVAVCTEIEKKTKKKLILDYLWENLRYHNWWAYRYYLCEALALVNVIGQMFLMNRFFDGEFMTFGLKVIKYMESDQEIRIDPMIYIFPRMVKCTLFNKFGSSGEVERHDALCILPLNVVNEKIYIFLWFWFVILGFLTFITLIYRIIIIFSPRMRVYMMRMRFRLVRRDNIDTIVRRSKMGDWYLLYILGENLDSIIFRDIMQEFANKLNHNYQHHIHGVPDA
Summary
Description
Structural component of the gap junctions.
Structural component of the gap junctions at electrical synapses in distal and mid-depth levels in the lamina.
Structural component of the gap junctions at electrical synapses in distal and mid-depth levels in the lamina. Isoform Lethal forms voltage sensitive intercellular channels through homotypic interactions.
Structural component of the gap junctions at electrical synapses in distal and mid-depth levels in the lamina.
Structural component of the gap junctions at electrical synapses in distal and mid-depth levels in the lamina. Isoform Lethal forms voltage sensitive intercellular channels through homotypic interactions.
Subunit
Monomer.
Monomer (isoform Lethal).
Monomer (isoform Lethal).
Similarity
Belongs to the pannexin family.
Keywords
Cell junction
Cell membrane
Complete proteome
Gap junction
Ion channel
Ion transport
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Alternative splicing
Behavior
Feature
chain Innexin shaking-B
splice variant In isoform E.
splice variant In isoform E.
Uniprot
A0A0N1IHM3
A0A437BXR1
A0A2A4K6A3
A0A194PPI7
A0A212EW68
A0A336LII3
+ More
A0A1I8PXW4 A0A182YB94 B4JXM9 B5DNE9 B4M3P9 B4L370 B4PYJ7 B4I6B1 B3NY39 Q1DH70 A0A1W4VVW7 B0XHF1 P33085 A0A2C9GPG3 A0A182L9C6 Q7PXN1 B4MSW8 A0A0A1XT70 A0A0K8WG78 A0A034WH33 W8B2W5 A0A1J1HVY9 A0A195ERQ0 A0A026WUY9 A0A1B6EBL0 A0A158NQK3 A0A0C9QHU7 A0A1W4XQJ8 J9JSD1 A0A2H8TIN1 A0A2S2PYJ4 A0A1Y1M793 K7J6Q6 B3N0Z8 A0A1B6G3H6 A0A1B6HU79 D6X4I2 E0VDS4 T1HVH6 E9G961 A0A1B6MLB6 A0A1I8PXX3 C0KLS8 N6U651 A0A0L0BU34 A0A1I8PXU9 A0A0Q9WMJ0 A0A0Q9WMI1 A0A0R3P0T5 A0A0Q9WDL3 A0A0Q9WM73 A0A3B0K1Z9 A0A3B0K037 A0A0R3P0X1 A0A1I8PXY8 A0A0R3P668 A0A0R1EC29 A0A0Q5T681 P33085-4 M9NGR8 A0A1B0GAK5 P33085-2 A0A1W4VIF9 G4LU30 A0A0K8U1E9 A0A195D3W6 A0A1W7R8S4 F5HMP1 A0A336LJS9 A0A0R3P573 A0A0Q9WP89 A0A226EJ08 A0A3R7N7V3 A0A068BKA8 A0A146LYX0 A0A097KUR2 A0A0K2V323 A0A1D2N4L7 A0A0A9X0I9 A0A2J7PH97 A0A2P6L614 A0A0J7KZE2 A0A3L8E3W4 A0A023F430 A0A154NVX6 A0A195BQZ2 F4WKA1 A0A088A2D6 T1JJS6 A0A0L7R3P8 A0A0Q5TJ67 A0A2A3EBF3 A0A131XRY4 E1ZXF2
A0A1I8PXW4 A0A182YB94 B4JXM9 B5DNE9 B4M3P9 B4L370 B4PYJ7 B4I6B1 B3NY39 Q1DH70 A0A1W4VVW7 B0XHF1 P33085 A0A2C9GPG3 A0A182L9C6 Q7PXN1 B4MSW8 A0A0A1XT70 A0A0K8WG78 A0A034WH33 W8B2W5 A0A1J1HVY9 A0A195ERQ0 A0A026WUY9 A0A1B6EBL0 A0A158NQK3 A0A0C9QHU7 A0A1W4XQJ8 J9JSD1 A0A2H8TIN1 A0A2S2PYJ4 A0A1Y1M793 K7J6Q6 B3N0Z8 A0A1B6G3H6 A0A1B6HU79 D6X4I2 E0VDS4 T1HVH6 E9G961 A0A1B6MLB6 A0A1I8PXX3 C0KLS8 N6U651 A0A0L0BU34 A0A1I8PXU9 A0A0Q9WMJ0 A0A0Q9WMI1 A0A0R3P0T5 A0A0Q9WDL3 A0A0Q9WM73 A0A3B0K1Z9 A0A3B0K037 A0A0R3P0X1 A0A1I8PXY8 A0A0R3P668 A0A0R1EC29 A0A0Q5T681 P33085-4 M9NGR8 A0A1B0GAK5 P33085-2 A0A1W4VIF9 G4LU30 A0A0K8U1E9 A0A195D3W6 A0A1W7R8S4 F5HMP1 A0A336LJS9 A0A0R3P573 A0A0Q9WP89 A0A226EJ08 A0A3R7N7V3 A0A068BKA8 A0A146LYX0 A0A097KUR2 A0A0K2V323 A0A1D2N4L7 A0A0A9X0I9 A0A2J7PH97 A0A2P6L614 A0A0J7KZE2 A0A3L8E3W4 A0A023F430 A0A154NVX6 A0A195BQZ2 F4WKA1 A0A088A2D6 T1JJS6 A0A0L7R3P8 A0A0Q5TJ67 A0A2A3EBF3 A0A131XRY4 E1ZXF2
Pubmed
26354079
22118469
25244985
17994087
15632085
17550304
+ More
17510324 1487155 8500183 7601305 7892218 10731132 12537572 9552170 9428764 10440730 20966253 12364791 25830018 25348373 24495485 24508170 21347285 28004739 20075255 18362917 19820115 20566863 21292972 23537049 26108605 18057021 23185243 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 14747013 17210077 26823975 25210156 27289101 25401762 30249741 25474469 21719571 20798317
17510324 1487155 8500183 7601305 7892218 10731132 12537572 9552170 9428764 10440730 20966253 12364791 25830018 25348373 24495485 24508170 21347285 28004739 20075255 18362917 19820115 20566863 21292972 23537049 26108605 18057021 23185243 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 14747013 17210077 26823975 25210156 27289101 25401762 30249741 25474469 21719571 20798317
EMBL
KQ460497
KPJ14316.1
RSAL01000001
RVE55228.1
NWSH01000135
PCG79200.1
+ More
KQ459596 KPI95356.1 AGBW02012071 OWR45726.1 UFQT01000021 SSX17914.1 CH916376 EDV95128.1 CH379064 EDY72957.1 CH940651 EDW65424.1 CH933810 EDW06998.1 CM000162 EDX03038.1 CH480823 EDW56317.1 CH954180 EDV47560.1 CH899795 EAT32503.1 DS233153 EDS28380.1 M98872 X65103 L13306 S78495 U17330 AE014298 BT025062 APCN01000876 AAAB01008987 EAA00824.1 CH963851 EDW75207.1 GBXI01000142 JAD14150.1 GDHF01002146 JAI50168.1 GAKP01005884 JAC53068.1 GAMC01018964 JAB87591.1 CVRI01000024 CRK92232.1 KQ982021 KYN30554.1 KK107087 EZA59865.1 GEDC01001987 JAS35311.1 ADTU01023365 ADTU01023366 GBYB01000132 JAG69899.1 ABLF02036239 GFXV01001323 MBW13128.1 GGMS01001412 MBY70615.1 GEZM01043354 JAV79177.1 AAZX01003113 CH902650 EDV30033.2 GECZ01012779 JAS56990.1 GECU01029505 GECU01007631 JAS78201.1 JAT00076.1 KQ971380 EEZ97266.1 AAZO01001464 DS235084 EEB11530.1 ACPB03015783 GL732535 EFX84089.1 GEBQ01003273 JAT36704.1 FJ619505 ACN41956.1 APGK01041017 KB740987 KB631964 ENN76126.1 ERL87521.1 JRES01001334 KNC23590.1 KRF93903.1 KRF82228.1 KRT06878.1 KRF82230.1 KRF93906.1 OUUW01000011 SPP86692.1 SPP86693.1 KRT06881.1 KRT06880.1 KRK07080.1 KQS30559.1 AFH07500.1 CCAG010019535 CCAG010019536 CCAG010019537 CCAG010019538 CCAG010019539 CCAG010019540 CCAG010019541 BT132660 AEQ05562.1 GDHF01032154 JAI20160.1 KQ976885 KYN07541.1 GEHC01000103 JAV47542.1 EGK97563.1 SSX17915.1 KRT06879.1 KRF82229.1 LNIX01000003 OXA57280.1 QCYY01001224 ROT79567.1 KJ642222 AID07491.1 GDHC01005838 JAQ12791.1 KM984501 AIT97136.1 HACA01027523 CDW44884.1 LJIJ01000239 ODN00005.1 GBHO01031301 JAG12303.1 NEVH01025142 PNF15698.1 MWRG01001558 PRD34022.1 LBMM01001661 KMQ95927.1 QOIP01000001 RLU26919.1 GBBI01002467 JAC16245.1 KQ434769 KZC03797.1 KQ976419 KYM89303.1 GL888199 EGI65325.1 JH431978 KQ414663 KOC65366.1 KQS30558.1 KZ288311 PBC28516.1 GEFM01006818 JAP68978.1 GL435038 EFN74129.1
KQ459596 KPI95356.1 AGBW02012071 OWR45726.1 UFQT01000021 SSX17914.1 CH916376 EDV95128.1 CH379064 EDY72957.1 CH940651 EDW65424.1 CH933810 EDW06998.1 CM000162 EDX03038.1 CH480823 EDW56317.1 CH954180 EDV47560.1 CH899795 EAT32503.1 DS233153 EDS28380.1 M98872 X65103 L13306 S78495 U17330 AE014298 BT025062 APCN01000876 AAAB01008987 EAA00824.1 CH963851 EDW75207.1 GBXI01000142 JAD14150.1 GDHF01002146 JAI50168.1 GAKP01005884 JAC53068.1 GAMC01018964 JAB87591.1 CVRI01000024 CRK92232.1 KQ982021 KYN30554.1 KK107087 EZA59865.1 GEDC01001987 JAS35311.1 ADTU01023365 ADTU01023366 GBYB01000132 JAG69899.1 ABLF02036239 GFXV01001323 MBW13128.1 GGMS01001412 MBY70615.1 GEZM01043354 JAV79177.1 AAZX01003113 CH902650 EDV30033.2 GECZ01012779 JAS56990.1 GECU01029505 GECU01007631 JAS78201.1 JAT00076.1 KQ971380 EEZ97266.1 AAZO01001464 DS235084 EEB11530.1 ACPB03015783 GL732535 EFX84089.1 GEBQ01003273 JAT36704.1 FJ619505 ACN41956.1 APGK01041017 KB740987 KB631964 ENN76126.1 ERL87521.1 JRES01001334 KNC23590.1 KRF93903.1 KRF82228.1 KRT06878.1 KRF82230.1 KRF93906.1 OUUW01000011 SPP86692.1 SPP86693.1 KRT06881.1 KRT06880.1 KRK07080.1 KQS30559.1 AFH07500.1 CCAG010019535 CCAG010019536 CCAG010019537 CCAG010019538 CCAG010019539 CCAG010019540 CCAG010019541 BT132660 AEQ05562.1 GDHF01032154 JAI20160.1 KQ976885 KYN07541.1 GEHC01000103 JAV47542.1 EGK97563.1 SSX17915.1 KRT06879.1 KRF82229.1 LNIX01000003 OXA57280.1 QCYY01001224 ROT79567.1 KJ642222 AID07491.1 GDHC01005838 JAQ12791.1 KM984501 AIT97136.1 HACA01027523 CDW44884.1 LJIJ01000239 ODN00005.1 GBHO01031301 JAG12303.1 NEVH01025142 PNF15698.1 MWRG01001558 PRD34022.1 LBMM01001661 KMQ95927.1 QOIP01000001 RLU26919.1 GBBI01002467 JAC16245.1 KQ434769 KZC03797.1 KQ976419 KYM89303.1 GL888199 EGI65325.1 JH431978 KQ414663 KOC65366.1 KQS30558.1 KZ288311 PBC28516.1 GEFM01006818 JAP68978.1 GL435038 EFN74129.1
Proteomes
UP000053240
UP000283053
UP000218220
UP000053268
UP000007151
UP000095300
+ More
UP000076408 UP000001070 UP000001819 UP000008792 UP000009192 UP000002282 UP000001292 UP000008711 UP000008820 UP000192221 UP000002320 UP000000803 UP000075840 UP000075882 UP000007062 UP000007798 UP000183832 UP000078541 UP000053097 UP000005205 UP000192223 UP000007819 UP000002358 UP000007801 UP000007266 UP000009046 UP000015103 UP000000305 UP000019118 UP000030742 UP000037069 UP000268350 UP000092444 UP000078542 UP000198287 UP000283509 UP000094527 UP000235965 UP000036403 UP000279307 UP000076502 UP000078540 UP000007755 UP000005203 UP000053825 UP000242457 UP000000311
UP000076408 UP000001070 UP000001819 UP000008792 UP000009192 UP000002282 UP000001292 UP000008711 UP000008820 UP000192221 UP000002320 UP000000803 UP000075840 UP000075882 UP000007062 UP000007798 UP000183832 UP000078541 UP000053097 UP000005205 UP000192223 UP000007819 UP000002358 UP000007801 UP000007266 UP000009046 UP000015103 UP000000305 UP000019118 UP000030742 UP000037069 UP000268350 UP000092444 UP000078542 UP000198287 UP000283509 UP000094527 UP000235965 UP000036403 UP000279307 UP000076502 UP000078540 UP000007755 UP000005203 UP000053825 UP000242457 UP000000311
Pfam
PF00876 Innexin
Interpro
IPR000990
Innexin
ProteinModelPortal
A0A0N1IHM3
A0A437BXR1
A0A2A4K6A3
A0A194PPI7
A0A212EW68
A0A336LII3
+ More
A0A1I8PXW4 A0A182YB94 B4JXM9 B5DNE9 B4M3P9 B4L370 B4PYJ7 B4I6B1 B3NY39 Q1DH70 A0A1W4VVW7 B0XHF1 P33085 A0A2C9GPG3 A0A182L9C6 Q7PXN1 B4MSW8 A0A0A1XT70 A0A0K8WG78 A0A034WH33 W8B2W5 A0A1J1HVY9 A0A195ERQ0 A0A026WUY9 A0A1B6EBL0 A0A158NQK3 A0A0C9QHU7 A0A1W4XQJ8 J9JSD1 A0A2H8TIN1 A0A2S2PYJ4 A0A1Y1M793 K7J6Q6 B3N0Z8 A0A1B6G3H6 A0A1B6HU79 D6X4I2 E0VDS4 T1HVH6 E9G961 A0A1B6MLB6 A0A1I8PXX3 C0KLS8 N6U651 A0A0L0BU34 A0A1I8PXU9 A0A0Q9WMJ0 A0A0Q9WMI1 A0A0R3P0T5 A0A0Q9WDL3 A0A0Q9WM73 A0A3B0K1Z9 A0A3B0K037 A0A0R3P0X1 A0A1I8PXY8 A0A0R3P668 A0A0R1EC29 A0A0Q5T681 P33085-4 M9NGR8 A0A1B0GAK5 P33085-2 A0A1W4VIF9 G4LU30 A0A0K8U1E9 A0A195D3W6 A0A1W7R8S4 F5HMP1 A0A336LJS9 A0A0R3P573 A0A0Q9WP89 A0A226EJ08 A0A3R7N7V3 A0A068BKA8 A0A146LYX0 A0A097KUR2 A0A0K2V323 A0A1D2N4L7 A0A0A9X0I9 A0A2J7PH97 A0A2P6L614 A0A0J7KZE2 A0A3L8E3W4 A0A023F430 A0A154NVX6 A0A195BQZ2 F4WKA1 A0A088A2D6 T1JJS6 A0A0L7R3P8 A0A0Q5TJ67 A0A2A3EBF3 A0A131XRY4 E1ZXF2
A0A1I8PXW4 A0A182YB94 B4JXM9 B5DNE9 B4M3P9 B4L370 B4PYJ7 B4I6B1 B3NY39 Q1DH70 A0A1W4VVW7 B0XHF1 P33085 A0A2C9GPG3 A0A182L9C6 Q7PXN1 B4MSW8 A0A0A1XT70 A0A0K8WG78 A0A034WH33 W8B2W5 A0A1J1HVY9 A0A195ERQ0 A0A026WUY9 A0A1B6EBL0 A0A158NQK3 A0A0C9QHU7 A0A1W4XQJ8 J9JSD1 A0A2H8TIN1 A0A2S2PYJ4 A0A1Y1M793 K7J6Q6 B3N0Z8 A0A1B6G3H6 A0A1B6HU79 D6X4I2 E0VDS4 T1HVH6 E9G961 A0A1B6MLB6 A0A1I8PXX3 C0KLS8 N6U651 A0A0L0BU34 A0A1I8PXU9 A0A0Q9WMJ0 A0A0Q9WMI1 A0A0R3P0T5 A0A0Q9WDL3 A0A0Q9WM73 A0A3B0K1Z9 A0A3B0K037 A0A0R3P0X1 A0A1I8PXY8 A0A0R3P668 A0A0R1EC29 A0A0Q5T681 P33085-4 M9NGR8 A0A1B0GAK5 P33085-2 A0A1W4VIF9 G4LU30 A0A0K8U1E9 A0A195D3W6 A0A1W7R8S4 F5HMP1 A0A336LJS9 A0A0R3P573 A0A0Q9WP89 A0A226EJ08 A0A3R7N7V3 A0A068BKA8 A0A146LYX0 A0A097KUR2 A0A0K2V323 A0A1D2N4L7 A0A0A9X0I9 A0A2J7PH97 A0A2P6L614 A0A0J7KZE2 A0A3L8E3W4 A0A023F430 A0A154NVX6 A0A195BQZ2 F4WKA1 A0A088A2D6 T1JJS6 A0A0L7R3P8 A0A0Q5TJ67 A0A2A3EBF3 A0A131XRY4 E1ZXF2
PDB
5H1R
E-value=2.72314e-15,
Score=199
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
Cell junction
Gap junction
Cell junction
Gap junction
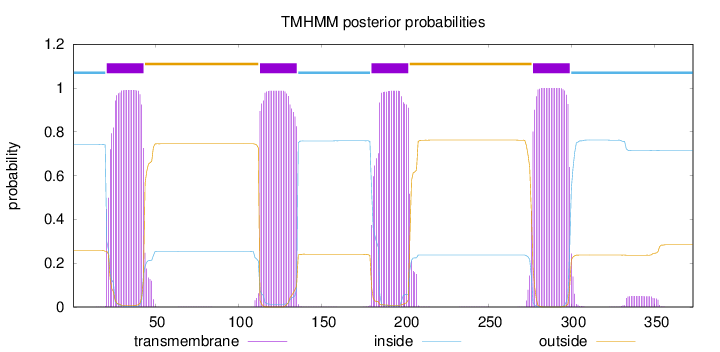
Length:
373
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
89.53429
Exp number, first 60 AAs:
21.49817
Total prob of N-in:
0.74194
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 112
TMhelix
113 - 135
inside
136 - 179
TMhelix
180 - 202
outside
203 - 276
TMhelix
277 - 299
inside
300 - 373
Population Genetic Test Statistics
Pi
249.759991
Theta
174.523726
Tajima's D
0.682071
CLR
0.709081
CSRT
0.567571621418929
Interpretation
Uncertain
