Gene
KWMTBOMO08623
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC105841899?_partial_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.672
Sequence
CDS
ATGAAGCCTGCAAAGGTTTTCAAAAACATCGCCAACGCGTTCAACACCTTGCCCTGGTCGGTGATCGCAGGGGCGCTGGAGTATCACGGCGTCCCTGCGTATCTCCGTCGGCTGATCGGGTCCTACCTCGAGGACAGGTCGGTCGTGTGCACCGGGCACGGTGGGGCGGTGCTCCGCTTCCCCGTCCAGCGCGGTGTTCCACAGGGGTCGGTCCTTGGCCCTCTCTTGTGGAACATCGGCTACGACTGGGTCTTGCGTGGCGCCCTAAGCACTCCTCTCCCGGGTCTGAGCGTAGTTTGCTACGCGGACGACACTTTGGTCGTGGCCCGGGGGAGGGACTTGCGAGAGTCTGCCCGTCTTTCCTGCGCGGGGGTGGCCTTCATCATCGGCAGGATCCGAAGGCTGGGTCTGGAGGTGGCGCTCGATAAATCCCAGGCCCTGTTGTTTCACGGGGCTCGGAGAGCGCCGCCTCAGGGGGCCCACCTCGTGATCGGAGGCGTTCGCGTCGAGATCGAGGCGACCGGGTTGCGGTACCTCGGTCTCGTACTGGACGGTCGTTGGAGCTTCCGCGCTCACTTTGAGAGATTAGGTCCCCGACTGATGGCGGCCGCCGGCTCGCTGAGTCGGCTGTTGCCGAACGTCGGGGGGCCTGACTTGGTGGTGCGCCGCCTCTACACGGGGGTGGTGCGGTCGATGGCACTGTACGGGGCTCCCGTGTGGTGCCACGCCCTGACCCGCGACAACATTGCGGCGTTGCGACGTCCGCAGCGCGCGATTGCGGTCAGGGTGGTTCGTGGATACCGCACCGTCTCGTTCGAGGCGGCGTGCGTGCTAGCTGGGACGCCTCCCTGGGACCTGGAAGCGGAGGCGCTCGCTGCGGATTACGCGTGGCGATGCGATCTCCGCTCCAGGGGGGAGCCGCGTCCTGGCGCGGCGGAAGTGCTCACGGGACACGGCTGTTTCGGTCGCTACCTGCACCTCGTCGCCCGGAGGGAGCCGACTCCGAAGTGCCACCACTGCAGTGGCTGCAACGAGGACACGGCGGAGCACACGCTCGCGTACTGCCCCACTGTCCTCGTTGCAAAAATAGGACCGGACTTGTCGCTTCCGACCGTCGTGGCTACGATGCTCGGCAGCGACGAGTCCTGGCAGGCGATGCTCGACTTCTGCGAGTCCACCATCTCGCAGAAGGAAGCGGCGGAAGGGAGAGCTCTTCTTCCCTCTCGGCGCCGTGCCGCCGCCGTCGGGCCGAGGGCCGGAGGAGGGCGTTTGTCCAGCTCCAGCCCCTATGAGGAGGCAGTCTCCTCCCGCGCGCTGGCACCAGGAGAACGGGACGGCGCGTCGGCGGCTGCGGACTGCGATGCGGTCCGCATTTTCGTCGTCGCTGTCGTCGCCGAACATACTGTTGAAGAGCAACCCGGTCGGCGGTGTATCGCGTTCCGACCCGGCAGGCTGGTTCTGGCCCAGCGGGGTATCCCGGAACACCAGCGGCACCGCCCGGGCGGCCTGGTAGGGCCGCCGTACCGCGGAGACTGA
Protein
MKPAKVFKNIANAFNTLPWSVIAGALEYHGVPAYLRRLIGSYLEDRSVVCTGHGGAVLRFPVQRGVPQGSVLGPLLWNIGYDWVLRGALSTPLPGLSVVCYADDTLVVARGRDLRESARLSCAGVAFIIGRIRRLGLEVALDKSQALLFHGARRAPPQGAHLVIGGVRVEIEATGLRYLGLVLDGRWSFRAHFERLGPRLMAAAGSLSRLLPNVGGPDLVVRRLYTGVVRSMALYGAPVWCHALTRDNIAALRRPQRAIAVRVVRGYRTVSFEAACVLAGTPPWDLEAEALAADYAWRCDLRSRGEPRPGAAEVLTGHGCFGRYLHLVARREPTPKCHHCSGCNEDTAEHTLAYCPTVLVAKIGPDLSLPTVVATMLGSDESWQAMLDFCESTISQKEAAEGRALLPSRRRAAAVGPRAGGGRLSSSSPYEEAVSSRALAPGERDGASAAADCDAVRIFVVAVVAEHTVEEQPGRRCIAFRPGRLVLAQRGIPEHQRHRPGGLVGPPYRGD
Summary
Uniprot
Q868Q4
A0A2W1B333
A0A0J7JZ13
A0A0J7JZF7
A0A0J7KIW1
A0A0J7KJG6
+ More
A0A0J7KLH8 A0A0J7KJH4 A0A0J7KF37 A0A0J7KYD2 A0A0J7KLT9 A0A0J7KIM6 A0A0J7KPZ3 A0A2H8TIT6 A0A0J7KCZ9 X1WMW4 J9LJ80 J9LVU7 A0A0J7MY17 A0A2H8TIE5 A0A2S2PEZ7 A0A0J7NIB5 J9JWJ3 A0A2M4BDI3 A0A2M4BBV9 X1XNG1 Q868S4 A0A2S2P488 A0A2M4BC01 D6WC19 F7IYV7 J9LBB9 T1DCM0 W8ANK7 W8ADE4 Q9N9Z1 A0A0J7KP02 F7IYV5
A0A0J7KLH8 A0A0J7KJH4 A0A0J7KF37 A0A0J7KYD2 A0A0J7KLT9 A0A0J7KIM6 A0A0J7KPZ3 A0A2H8TIT6 A0A0J7KCZ9 X1WMW4 J9LJ80 J9LVU7 A0A0J7MY17 A0A2H8TIE5 A0A2S2PEZ7 A0A0J7NIB5 J9JWJ3 A0A2M4BDI3 A0A2M4BBV9 X1XNG1 Q868S4 A0A2S2P488 A0A2M4BC01 D6WC19 F7IYV7 J9LBB9 T1DCM0 W8ANK7 W8ADE4 Q9N9Z1 A0A0J7KP02 F7IYV5
EMBL
AB090825
BAC57926.1
KZ150386
PZC71062.1
LBMM01019839
KMQ83438.1
+ More
LBMM01019838 KMQ83439.1 LBMM01006725 KMQ90373.1 LBMM01006671 KMQ90407.1 LBMM01005948 KMQ91066.1 LBMM01006657 KMQ90417.1 LBMM01008448 KMQ88867.1 LBMM01002052 KMQ95336.1 LBMM01005733 KMQ91252.1 LBMM01007036 KMQ90099.1 LBMM01004505 KMQ92346.1 GFXV01002242 MBW14047.1 LBMM01009527 LBMM01009515 KMQ88059.1 KMQ88067.1 ABLF02059310 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 ABLF02034925 LBMM01014122 KMQ85315.1 GFXV01002090 MBW13895.1 GGMR01015159 MBY27778.1 LBMM01004603 KMQ92250.1 ABLF02013358 ABLF02013361 ABLF02054869 GGFJ01001727 MBW50868.1 GGFJ01001369 MBW50510.1 ABLF02043108 AB090815 BAC57906.1 GGMR01011682 MBY24301.1 GGFJ01001370 MBW50511.1 KQ971309 EEZ99146.1 AB593327 BAK38648.1 ABLF02020562 GALA01001777 JAA93075.1 GAMC01020517 JAB86038.1 GAMC01020519 GAMC01020515 JAB86040.1 AJ278684 CAB99192.1 LBMM01004906 KMQ92001.1 AB593325 BAK38646.1
LBMM01019838 KMQ83439.1 LBMM01006725 KMQ90373.1 LBMM01006671 KMQ90407.1 LBMM01005948 KMQ91066.1 LBMM01006657 KMQ90417.1 LBMM01008448 KMQ88867.1 LBMM01002052 KMQ95336.1 LBMM01005733 KMQ91252.1 LBMM01007036 KMQ90099.1 LBMM01004505 KMQ92346.1 GFXV01002242 MBW14047.1 LBMM01009527 LBMM01009515 KMQ88059.1 KMQ88067.1 ABLF02059310 ABLF02015311 ABLF02015320 ABLF02034009 ABLF02043484 ABLF02034925 LBMM01014122 KMQ85315.1 GFXV01002090 MBW13895.1 GGMR01015159 MBY27778.1 LBMM01004603 KMQ92250.1 ABLF02013358 ABLF02013361 ABLF02054869 GGFJ01001727 MBW50868.1 GGFJ01001369 MBW50510.1 ABLF02043108 AB090815 BAC57906.1 GGMR01011682 MBY24301.1 GGFJ01001370 MBW50511.1 KQ971309 EEZ99146.1 AB593327 BAK38648.1 ABLF02020562 GALA01001777 JAA93075.1 GAMC01020517 JAB86038.1 GAMC01020519 GAMC01020515 JAB86040.1 AJ278684 CAB99192.1 LBMM01004906 KMQ92001.1 AB593325 BAK38646.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
Q868Q4
A0A2W1B333
A0A0J7JZ13
A0A0J7JZF7
A0A0J7KIW1
A0A0J7KJG6
+ More
A0A0J7KLH8 A0A0J7KJH4 A0A0J7KF37 A0A0J7KYD2 A0A0J7KLT9 A0A0J7KIM6 A0A0J7KPZ3 A0A2H8TIT6 A0A0J7KCZ9 X1WMW4 J9LJ80 J9LVU7 A0A0J7MY17 A0A2H8TIE5 A0A2S2PEZ7 A0A0J7NIB5 J9JWJ3 A0A2M4BDI3 A0A2M4BBV9 X1XNG1 Q868S4 A0A2S2P488 A0A2M4BC01 D6WC19 F7IYV7 J9LBB9 T1DCM0 W8ANK7 W8ADE4 Q9N9Z1 A0A0J7KP02 F7IYV5
A0A0J7KLH8 A0A0J7KJH4 A0A0J7KF37 A0A0J7KYD2 A0A0J7KLT9 A0A0J7KIM6 A0A0J7KPZ3 A0A2H8TIT6 A0A0J7KCZ9 X1WMW4 J9LJ80 J9LVU7 A0A0J7MY17 A0A2H8TIE5 A0A2S2PEZ7 A0A0J7NIB5 J9JWJ3 A0A2M4BDI3 A0A2M4BBV9 X1XNG1 Q868S4 A0A2S2P488 A0A2M4BC01 D6WC19 F7IYV7 J9LBB9 T1DCM0 W8ANK7 W8ADE4 Q9N9Z1 A0A0J7KP02 F7IYV5
Ontologies
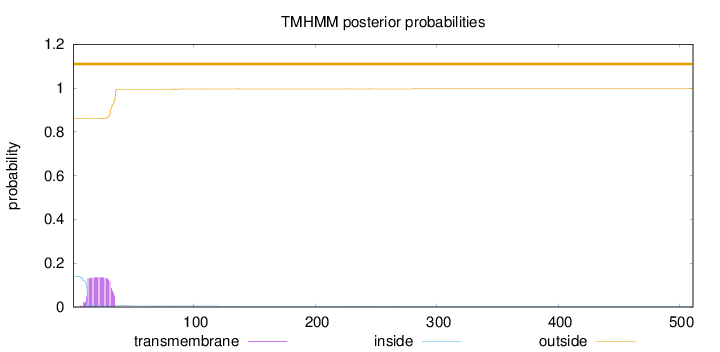
Topology
Length:
511
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.96775000000001
Exp number, first 60 AAs:
2.91983
Total prob of N-in:
0.13969
outside
1 - 511
Population Genetic Test Statistics
Pi
0.204735
Theta
1.857583
Tajima's D
-2.064798
CLR
14.428757
CSRT
0.00419979001049947
Interpretation
Possibly Positive selection
