Gene
KWMTBOMO08622
Annotation
Innexin_shaking-B_[Papilio_xuthus]
Full name
Innexin
+ More
Innexin shaking-B
Innexin shaking-B
Alternative Name
Protein passover
Location in the cell
Extracellular Reliability : 2.102
Sequence
CDS
ATGGAGATCCTCACCAGCATCTACGCGCTGCTGTCTGGAGGGCAAGGTGTTAATCCGAAGATCGACTCGGTAGCATTTCGTCTGCACTGCGGGGCAACAACTGCCGCGTTGTTGGCGGCGAGCGCGGCTCTCACCACGCGCCACCTAGTGGGCAACCCGATAGACTGCATCCACACAAGGCAAGCTACACTGCACGCTAAGTCACCTGTACTTTGTTCATAA
Protein
MEILTSIYALLSGGQGVNPKIDSVAFRLHCGATTAALLAASAALTTRHLVGNPIDCIHTRQATLHAKSPVLCS
Summary
Description
Structural component of the gap junctions.
Structural component of the gap junctions at electrical synapses in distal and mid-depth levels in the lamina. Isoform Lethal forms voltage sensitive intercellular channels through homotypic interactions.
Structural component of the gap junctions at electrical synapses in distal and mid-depth levels in the lamina. Isoform Lethal forms voltage sensitive intercellular channels through homotypic interactions.
Subunit
Monomer (isoform Lethal).
Similarity
Belongs to the pannexin family.
Keywords
Alternative splicing
Behavior
Cell junction
Cell membrane
Complete proteome
Gap junction
Ion channel
Ion transport
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Innexin shaking-B
splice variant In isoform E.
splice variant In isoform E.
Uniprot
A0A2H1WLU8
A0A194PW45
A0A212EW91
A0A2A4K556
A0A0N1PI77
A0A1I8PXX3
+ More
A0A336LJS9 A0A336LII3 A0A1B0CA92 A0A0Q5TJ67 A0A0Q9WDL3 A0A3B0K037 A0A0K8U1E9 G4LU30 A0A0R1EC29 A0A0L0BU34 A0A0Q5T681 A0A1W4VIF9 A0A0Q9WM73 A0A0R3P0X1 M9NGR8 A0A1I8PXY8 A0A1I8PXU9 A0A0Q9WMI1 A0A0Q9WMJ0 A0A0R3P0T5 A0A3B0K1Z9 P33085-4 A0A1W4XEX8 A0A182PGZ8 A0A2P8XJM0 W5JKZ7 A0A182R507 A0A1W4XQL8 A0A2P8XJL9 D6X4I4 A0A182PZH7
A0A336LJS9 A0A336LII3 A0A1B0CA92 A0A0Q5TJ67 A0A0Q9WDL3 A0A3B0K037 A0A0K8U1E9 G4LU30 A0A0R1EC29 A0A0L0BU34 A0A0Q5T681 A0A1W4VIF9 A0A0Q9WM73 A0A0R3P0X1 M9NGR8 A0A1I8PXY8 A0A1I8PXU9 A0A0Q9WMI1 A0A0Q9WMJ0 A0A0R3P0T5 A0A3B0K1Z9 P33085-4 A0A1W4XEX8 A0A182PGZ8 A0A2P8XJM0 W5JKZ7 A0A182R507 A0A1W4XQL8 A0A2P8XJL9 D6X4I4 A0A182PZH7
Pubmed
EMBL
ODYU01009547
SOQ54051.1
KQ459596
KPI95355.1
AGBW02012071
OWR45727.1
+ More
NWSH01000135 PCG79199.1 KQ460497 KPJ14314.1 UFQT01000021 SSX17915.1 SSX17914.1 AJWK01003433 CH954180 KQS30558.1 CH940651 KRF82230.1 OUUW01000011 SPP86693.1 GDHF01032154 JAI20160.1 BT132660 AEQ05562.1 CM000162 KRK07080.1 JRES01001334 KNC23590.1 KQS30559.1 CH933810 KRF93906.1 CH379064 KRT06881.1 AE014298 AFH07500.1 KRF82228.1 KRF93903.1 KRT06878.1 SPP86692.1 M98872 X65103 L13306 S78495 U17330 BT025062 PYGN01001910 PSN32196.1 ADMH02001137 ETN63973.1 PSN32195.1 KQ971380 EEZ97265.1 AXCN02001523 AXCN02001524
NWSH01000135 PCG79199.1 KQ460497 KPJ14314.1 UFQT01000021 SSX17915.1 SSX17914.1 AJWK01003433 CH954180 KQS30558.1 CH940651 KRF82230.1 OUUW01000011 SPP86693.1 GDHF01032154 JAI20160.1 BT132660 AEQ05562.1 CM000162 KRK07080.1 JRES01001334 KNC23590.1 KQS30559.1 CH933810 KRF93906.1 CH379064 KRT06881.1 AE014298 AFH07500.1 KRF82228.1 KRF93903.1 KRT06878.1 SPP86692.1 M98872 X65103 L13306 S78495 U17330 BT025062 PYGN01001910 PSN32196.1 ADMH02001137 ETN63973.1 PSN32195.1 KQ971380 EEZ97265.1 AXCN02001523 AXCN02001524
Proteomes
Pfam
PF00876 Innexin
Interpro
IPR000990
Innexin
ProteinModelPortal
A0A2H1WLU8
A0A194PW45
A0A212EW91
A0A2A4K556
A0A0N1PI77
A0A1I8PXX3
+ More
A0A336LJS9 A0A336LII3 A0A1B0CA92 A0A0Q5TJ67 A0A0Q9WDL3 A0A3B0K037 A0A0K8U1E9 G4LU30 A0A0R1EC29 A0A0L0BU34 A0A0Q5T681 A0A1W4VIF9 A0A0Q9WM73 A0A0R3P0X1 M9NGR8 A0A1I8PXY8 A0A1I8PXU9 A0A0Q9WMI1 A0A0Q9WMJ0 A0A0R3P0T5 A0A3B0K1Z9 P33085-4 A0A1W4XEX8 A0A182PGZ8 A0A2P8XJM0 W5JKZ7 A0A182R507 A0A1W4XQL8 A0A2P8XJL9 D6X4I4 A0A182PZH7
A0A336LJS9 A0A336LII3 A0A1B0CA92 A0A0Q5TJ67 A0A0Q9WDL3 A0A3B0K037 A0A0K8U1E9 G4LU30 A0A0R1EC29 A0A0L0BU34 A0A0Q5T681 A0A1W4VIF9 A0A0Q9WM73 A0A0R3P0X1 M9NGR8 A0A1I8PXY8 A0A1I8PXU9 A0A0Q9WMI1 A0A0Q9WMJ0 A0A0R3P0T5 A0A3B0K1Z9 P33085-4 A0A1W4XEX8 A0A182PGZ8 A0A2P8XJM0 W5JKZ7 A0A182R507 A0A1W4XQL8 A0A2P8XJL9 D6X4I4 A0A182PZH7
Ontologies
KEGG
GO
PANTHER
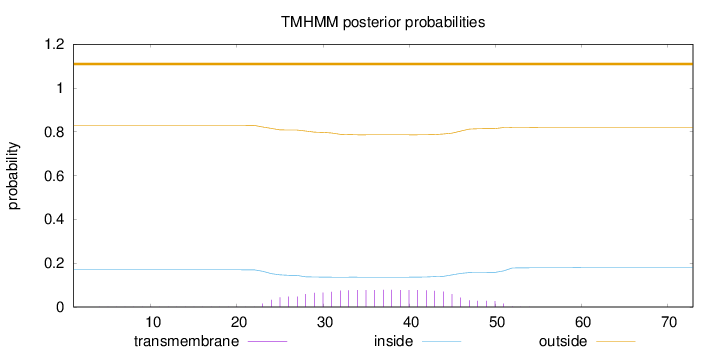
Topology
Subcellular location
Cell membrane
Cell junction
Gap junction
Cell junction
Gap junction
Length:
73
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.67605
Exp number, first 60 AAs:
1.67605
Total prob of N-in:
0.17029
outside
1 - 73
Population Genetic Test Statistics
Pi
190.384806
Theta
186.692038
Tajima's D
0.17453
CLR
0
CSRT
0.417879106044698
Interpretation
Uncertain
