Gene
KWMTBOMO08617
Pre Gene Modal
BGIBMGA007621
Annotation
PREDICTED:_cap-specific_mRNA_(nucleoside-2'-O-)-methyltransferase_2_[Papilio_xuthus]
Full name
Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 2
Alternative Name
Cap methyltransferase 2 homolog
Cap2 2'O-ribose methyltransferase 2 homolog
FtsJ methyltransferase domain-containing protein 1 homolog
Protein Adrift
Cap2 2'O-ribose methyltransferase 2 homolog
FtsJ methyltransferase domain-containing protein 1 homolog
Protein Adrift
Location in the cell
Nuclear Reliability : 1.704 PlasmaMembrane Reliability : 1.563
Sequence
CDS
ATGGACGGATCAATAAAAAAAGATGATGTAAAATCTGAGCGTAAGATGGTATCATTGCACTTGTGTGAGGCACCGGGAGCATTCATCACATCTTTGAACCACTATTTGAAGTTAAATCATCCTGGGATACAGTGGACCTGGGTTGCTAACACACTAAATCCATACTATGAGGGTAATTCCCCATCGAGCATGATCAGTGATGACAGGTTCATGTTTCACACCCTAGATAACTGGCATTTTGGGGTAGACAATACTGGTAACTTAATGGATTGGGAAAATTCACAAGCCATCATTAGAAAAGCGAAATCCCTTGGAAAGGTTATGTTAGTGACGGCAGATGGCTCCATTGATTGTCTACAGGCACCTGATGCACAGGAAGAGGCAACATCTCTATTACATTACTGTGAAATCACTACTGCCCTGCAAGCATTAAGCCAGGGTGGAACTTTAATATTCAAGCTTTTTACTATCTTCGAACATAGCACTGTGAGTCTGCTTTACCTAATTAATCATTTGTTCTCTGAAGTTAACATCTACAAACCGGTCACCTCGAGACAGGGCAACTCTGAAGTGTATGCAGTGTGCTTAAATTATAAAGGAATGTCTATACTGGAGAAGTTTACGCCGGTACTCATGTCAGCATATGGAACGGAACTATATGGAAATTTATCATTATTCCGACTTGATGACATCCCAGATACATTTTTGAAGCAAATCCAAGAGTGCGCATGTTACTTTTGTTCAATACAATGTCAAGTTATCAATAATAACCTGCAGGCATACTTAATGCAGAATAATATCGCTCTACACCGAAATATAAAGAAAATAAGGGCTGTTGTTGCTGACGAATTTATTTGGCATTATGACTTGAAACCCTTAAATTCTCATGAGGAGATACTCAAAGGTATTCTACATGAGGAGAACAAAATCAATATGAATCCCAGATACCACCGAGGATCTTACACTGAACGACAGCTCTATTCAAAGATGTCTTTACGAGAGAAATTGCACAATTTGAACTCATATCTGCATGCTGAAGTCCTAACGAATCCTACAGTGTTGATAAATGAACCGGTCAAATGGAAGAGATTTTATGATGCTCCTAAAGAAATAGAATTAACATTTACTTTTGGACGACCTTTACAAAAAATTAATAGCTCAAAATTTATATTCGTGCCAATATTTAAGCTATATCAAAAGATTTTAGAAGAGAAAGAATTTAAACACATTGCAGTATGTACAGATAGTAATCGTGATGTCAATTTAACGGCACTAGATTATGAAGTGGACAAAGTGCTCTGCGTACAGGAGTACCGTTGCAGTGAAAGTTTTTATAAATATGAGAAAAACTGTTTTAGATCTATAATGAAATATTTGAGAAATTTATCTTCAAACGAGACTCTTGTAATACAGAATATGAATGCTTTAACCCATTTTATCGTGAGCATTCTATATATTCTCTCTGTTAAATGTTTCGAGAAAATTGGGTTTACATCAACCAATGGTATAGTTTTAAGAAATCTTAAGGATCCATCTGGACTGAAATACTTAGAAAGTATTGAAACTGAATGTGATAATGTATTAGATGATGACAAAAAGGATTTGTTGAATTCGTTGAATGTACAAACAACGAATGCCGGAGATTTTTACTATAGCATCGTGTTCTACAATAACACCTTTTACAGGAATAAGTGTATTGAGTATTTGTCAAAAATAGAGAAATTAATATAA
Protein
MDGSIKKDDVKSERKMVSLHLCEAPGAFITSLNHYLKLNHPGIQWTWVANTLNPYYEGNSPSSMISDDRFMFHTLDNWHFGVDNTGNLMDWENSQAIIRKAKSLGKVMLVTADGSIDCLQAPDAQEEATSLLHYCEITTALQALSQGGTLIFKLFTIFEHSTVSLLYLINHLFSEVNIYKPVTSRQGNSEVYAVCLNYKGMSILEKFTPVLMSAYGTELYGNLSLFRLDDIPDTFLKQIQECACYFCSIQCQVINNNLQAYLMQNNIALHRNIKKIRAVVADEFIWHYDLKPLNSHEEILKGILHEENKINMNPRYHRGSYTERQLYSKMSLREKLHNLNSYLHAEVLTNPTVLINEPVKWKRFYDAPKEIELTFTFGRPLQKINSSKFIFVPIFKLYQKILEEKEFKHIAVCTDSNRDVNLTALDYEVDKVLCVQEYRCSESFYKYEKNCFRSIMKYLRNLSSNETLVIQNMNALTHFIVSILYILSVKCFEKIGFTSTNGIVLRNLKDPSGLKYLESIETECDNVLDDDKKDLLNSLNVQTTNAGDFYYSIVFYNNTFYRNKCIEYLSKIEKLI
Summary
Description
Probable S-adenosyl-L-methionine-dependent methyltransferase that mediates mRNA cap2 2'-O-ribose methylation to the 5'-cap structure of mRNAs. May methylate the ribose of the second nucleotide of a m(7)GpppG-capped mRNA (cap0) to produce m(7)GpppRmpNm (cap2) (By similarity). Regulates expression of tracheal genes required for pathfinding on the segmental nerve (PubMed:10068643).
Catalytic Activity
a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-purine-ribonucleotide)-(ribonucleotide)-[mRNA] + S-adenosyl-L-methionine = a 5'-(N(7)-methyl 5'-triphosphoguanosine)-(2'-O-methyl-purine-ribonucleotide)-(2'-O-methyl-ribonucleotide)-[mRNA] + H(+) + S-adenosyl-L-homocysteine
Keywords
Complete proteome
Methyltransferase
mRNA capping
mRNA processing
Nucleus
Reference proteome
S-adenosyl-L-methionine
Transferase
Feature
chain Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 2
Uniprot
H9JDM4
A0A2H1W7T6
A0A2A4J9U8
A0A437AV96
A0A0N1IH66
A0A194PW40
+ More
A0A1B3PD74 A0A067QWQ8 A0A336LHW8 A0A182PL76 A0A084WDA9 A0A182Y0Q9 K7IXC0 A0A182T9P5 A0A182FHP5 A0A1Q3G139 A0A2P8ZED2 E0VPC7 Q174Y1 A0A182HM95 A0A182XCW6 Q7Q7A5 A0A2M4AM29 A0A336M145 A0A182UQT5 A0A232EI20 A0A182JZC2 A0A182QG34 A0A182MR21 A0A182WLT5 A0A182LJU1 W5JIT7 A0A182TN86 A0A182IL64 A0A2M4BER6 A0A151K098 A0A182NJY7 A0A1B3PDM5 A0A195BRL5 A0A151XIA7 A0A182RC32 A0A158NF86 F4WYN2 A0A3L8E3Y6 A0A1B6HI99 A0A151J8H2 A0A026WD51 A0A1I8PKZ7 A0A0L7QLK5 E2A158 A0A146M402 E9IAA2 A0A0V0G5M1 A0A1B0AMJ3 A0A1A9XH99 A0A224XJN1 T1PPH3 A0A0L0BPS5 A0A069DZE6 A0A1I8NDE1 A0A1B0AFR8 A0A1A9URB5 A0A1B0G1V7 A0A444T1Y3 A0A1B6BY81 Q9UAS6 S5G1K6 A0A091NBQ8 D6X511 A0A218V6L0 B4QBR4 A0A401Q195 I3MSR2 A0A2Y9RSW6 B4HN83 A0A1A9WYW9 A0A3B0J5B3 A0A0A1WJY9 H0Z6G6
A0A1B3PD74 A0A067QWQ8 A0A336LHW8 A0A182PL76 A0A084WDA9 A0A182Y0Q9 K7IXC0 A0A182T9P5 A0A182FHP5 A0A1Q3G139 A0A2P8ZED2 E0VPC7 Q174Y1 A0A182HM95 A0A182XCW6 Q7Q7A5 A0A2M4AM29 A0A336M145 A0A182UQT5 A0A232EI20 A0A182JZC2 A0A182QG34 A0A182MR21 A0A182WLT5 A0A182LJU1 W5JIT7 A0A182TN86 A0A182IL64 A0A2M4BER6 A0A151K098 A0A182NJY7 A0A1B3PDM5 A0A195BRL5 A0A151XIA7 A0A182RC32 A0A158NF86 F4WYN2 A0A3L8E3Y6 A0A1B6HI99 A0A151J8H2 A0A026WD51 A0A1I8PKZ7 A0A0L7QLK5 E2A158 A0A146M402 E9IAA2 A0A0V0G5M1 A0A1B0AMJ3 A0A1A9XH99 A0A224XJN1 T1PPH3 A0A0L0BPS5 A0A069DZE6 A0A1I8NDE1 A0A1B0AFR8 A0A1A9URB5 A0A1B0G1V7 A0A444T1Y3 A0A1B6BY81 Q9UAS6 S5G1K6 A0A091NBQ8 D6X511 A0A218V6L0 B4QBR4 A0A401Q195 I3MSR2 A0A2Y9RSW6 B4HN83 A0A1A9WYW9 A0A3B0J5B3 A0A0A1WJY9 H0Z6G6
EC Number
2.1.1.296
Pubmed
19121390
26354079
24845553
24438588
25244985
20075255
+ More
29403074 20566863 17510324 12364791 14747013 17210077 28648823 20966253 20920257 23761445 21347285 21719571 30249741 24508170 20798317 26823975 21282665 26108605 26334808 25315136 10068643 10731132 12537572 10731137 12537569 18362917 19820115 17994087 22936249 30297745 25830018 20360741
29403074 20566863 17510324 12364791 14747013 17210077 28648823 20966253 20920257 23761445 21347285 21719571 30249741 24508170 20798317 26823975 21282665 26108605 26334808 25315136 10068643 10731132 12537572 10731137 12537569 18362917 19820115 17994087 22936249 30297745 25830018 20360741
EMBL
BABH01003300
BABH01003301
ODYU01006885
SOQ49130.1
NWSH01002297
PCG68619.1
+ More
RSAL01000379 RVE42067.1 KQ459717 KPJ20281.1 KQ459596 KPI95350.1 KU659876 AOG17675.1 KK853274 KDR09094.1 UFQS01002149 UFQT01002149 SSX13342.1 SSX32776.1 ATLV01022979 KE525339 KFB48203.1 AAZX01013363 GFDL01001530 JAV33515.1 PYGN01000082 PSN54810.1 DS235363 EEB15233.1 CH477404 EAT41650.1 APCN01004381 AAAB01008960 EAA11922.4 GGFK01008486 MBW41807.1 UFQT01000305 SSX23051.1 NNAY01004368 OXU17995.1 AXCN02000491 AXCM01001849 ADMH02001054 ETN64292.1 GGFJ01002393 MBW51534.1 KQ981319 KYN42730.1 KU660000 AOG17798.1 KQ976417 KYM89655.1 KQ982080 KYQ60142.1 ADTU01014037 GL888454 EGI60695.1 QOIP01000001 RLU27367.1 GECU01033355 JAS74351.1 KQ979533 KYN21119.1 KK107260 EZA54000.1 KQ414924 KOC59396.1 GL435707 EFN72825.1 GDHC01004634 GDHC01002522 JAQ13995.1 JAQ16107.1 GL761997 EFZ22498.1 GECL01002853 JAP03271.1 JXJN01000465 GFTR01007624 JAW08802.1 KA650050 AFP64679.1 JRES01001644 KNC21224.1 GBGD01000775 JAC88114.1 CCAG010001059 SAUD01015087 RXG59651.1 GEDC01031090 JAS06208.1 AF117649 AE013599 AL035311 AY050236 BT150145 AGQ54568.1 KK844412 KFP86289.1 KQ971381 EEZ97212.2 MUZQ01000037 OWK61707.1 CM000362 CM002911 EDX07574.1 KMY94686.1 BFAA01011947 GCB79199.1 AGTP01010066 CH480816 EDW48365.1 OUUW01000001 SPP74913.1 GBXI01015574 JAC98717.1 ABQF01035986
RSAL01000379 RVE42067.1 KQ459717 KPJ20281.1 KQ459596 KPI95350.1 KU659876 AOG17675.1 KK853274 KDR09094.1 UFQS01002149 UFQT01002149 SSX13342.1 SSX32776.1 ATLV01022979 KE525339 KFB48203.1 AAZX01013363 GFDL01001530 JAV33515.1 PYGN01000082 PSN54810.1 DS235363 EEB15233.1 CH477404 EAT41650.1 APCN01004381 AAAB01008960 EAA11922.4 GGFK01008486 MBW41807.1 UFQT01000305 SSX23051.1 NNAY01004368 OXU17995.1 AXCN02000491 AXCM01001849 ADMH02001054 ETN64292.1 GGFJ01002393 MBW51534.1 KQ981319 KYN42730.1 KU660000 AOG17798.1 KQ976417 KYM89655.1 KQ982080 KYQ60142.1 ADTU01014037 GL888454 EGI60695.1 QOIP01000001 RLU27367.1 GECU01033355 JAS74351.1 KQ979533 KYN21119.1 KK107260 EZA54000.1 KQ414924 KOC59396.1 GL435707 EFN72825.1 GDHC01004634 GDHC01002522 JAQ13995.1 JAQ16107.1 GL761997 EFZ22498.1 GECL01002853 JAP03271.1 JXJN01000465 GFTR01007624 JAW08802.1 KA650050 AFP64679.1 JRES01001644 KNC21224.1 GBGD01000775 JAC88114.1 CCAG010001059 SAUD01015087 RXG59651.1 GEDC01031090 JAS06208.1 AF117649 AE013599 AL035311 AY050236 BT150145 AGQ54568.1 KK844412 KFP86289.1 KQ971381 EEZ97212.2 MUZQ01000037 OWK61707.1 CM000362 CM002911 EDX07574.1 KMY94686.1 BFAA01011947 GCB79199.1 AGTP01010066 CH480816 EDW48365.1 OUUW01000001 SPP74913.1 GBXI01015574 JAC98717.1 ABQF01035986
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000027135
+ More
UP000075885 UP000030765 UP000076408 UP000002358 UP000075901 UP000069272 UP000245037 UP000009046 UP000008820 UP000075840 UP000076407 UP000007062 UP000075903 UP000215335 UP000075881 UP000075886 UP000075883 UP000075920 UP000075882 UP000000673 UP000075902 UP000075880 UP000078541 UP000075884 UP000078540 UP000075809 UP000075900 UP000005205 UP000007755 UP000279307 UP000078492 UP000053097 UP000095300 UP000053825 UP000000311 UP000092460 UP000092443 UP000037069 UP000095301 UP000092445 UP000078200 UP000092444 UP000288706 UP000000803 UP000007266 UP000197619 UP000000304 UP000288216 UP000005215 UP000248480 UP000001292 UP000091820 UP000268350 UP000007754
UP000075885 UP000030765 UP000076408 UP000002358 UP000075901 UP000069272 UP000245037 UP000009046 UP000008820 UP000075840 UP000076407 UP000007062 UP000075903 UP000215335 UP000075881 UP000075886 UP000075883 UP000075920 UP000075882 UP000000673 UP000075902 UP000075880 UP000078541 UP000075884 UP000078540 UP000075809 UP000075900 UP000005205 UP000007755 UP000279307 UP000078492 UP000053097 UP000095300 UP000053825 UP000000311 UP000092460 UP000092443 UP000037069 UP000095301 UP000092445 UP000078200 UP000092444 UP000288706 UP000000803 UP000007266 UP000197619 UP000000304 UP000288216 UP000005215 UP000248480 UP000001292 UP000091820 UP000268350 UP000007754
Interpro
Gene 3D
ProteinModelPortal
H9JDM4
A0A2H1W7T6
A0A2A4J9U8
A0A437AV96
A0A0N1IH66
A0A194PW40
+ More
A0A1B3PD74 A0A067QWQ8 A0A336LHW8 A0A182PL76 A0A084WDA9 A0A182Y0Q9 K7IXC0 A0A182T9P5 A0A182FHP5 A0A1Q3G139 A0A2P8ZED2 E0VPC7 Q174Y1 A0A182HM95 A0A182XCW6 Q7Q7A5 A0A2M4AM29 A0A336M145 A0A182UQT5 A0A232EI20 A0A182JZC2 A0A182QG34 A0A182MR21 A0A182WLT5 A0A182LJU1 W5JIT7 A0A182TN86 A0A182IL64 A0A2M4BER6 A0A151K098 A0A182NJY7 A0A1B3PDM5 A0A195BRL5 A0A151XIA7 A0A182RC32 A0A158NF86 F4WYN2 A0A3L8E3Y6 A0A1B6HI99 A0A151J8H2 A0A026WD51 A0A1I8PKZ7 A0A0L7QLK5 E2A158 A0A146M402 E9IAA2 A0A0V0G5M1 A0A1B0AMJ3 A0A1A9XH99 A0A224XJN1 T1PPH3 A0A0L0BPS5 A0A069DZE6 A0A1I8NDE1 A0A1B0AFR8 A0A1A9URB5 A0A1B0G1V7 A0A444T1Y3 A0A1B6BY81 Q9UAS6 S5G1K6 A0A091NBQ8 D6X511 A0A218V6L0 B4QBR4 A0A401Q195 I3MSR2 A0A2Y9RSW6 B4HN83 A0A1A9WYW9 A0A3B0J5B3 A0A0A1WJY9 H0Z6G6
A0A1B3PD74 A0A067QWQ8 A0A336LHW8 A0A182PL76 A0A084WDA9 A0A182Y0Q9 K7IXC0 A0A182T9P5 A0A182FHP5 A0A1Q3G139 A0A2P8ZED2 E0VPC7 Q174Y1 A0A182HM95 A0A182XCW6 Q7Q7A5 A0A2M4AM29 A0A336M145 A0A182UQT5 A0A232EI20 A0A182JZC2 A0A182QG34 A0A182MR21 A0A182WLT5 A0A182LJU1 W5JIT7 A0A182TN86 A0A182IL64 A0A2M4BER6 A0A151K098 A0A182NJY7 A0A1B3PDM5 A0A195BRL5 A0A151XIA7 A0A182RC32 A0A158NF86 F4WYN2 A0A3L8E3Y6 A0A1B6HI99 A0A151J8H2 A0A026WD51 A0A1I8PKZ7 A0A0L7QLK5 E2A158 A0A146M402 E9IAA2 A0A0V0G5M1 A0A1B0AMJ3 A0A1A9XH99 A0A224XJN1 T1PPH3 A0A0L0BPS5 A0A069DZE6 A0A1I8NDE1 A0A1B0AFR8 A0A1A9URB5 A0A1B0G1V7 A0A444T1Y3 A0A1B6BY81 Q9UAS6 S5G1K6 A0A091NBQ8 D6X511 A0A218V6L0 B4QBR4 A0A401Q195 I3MSR2 A0A2Y9RSW6 B4HN83 A0A1A9WYW9 A0A3B0J5B3 A0A0A1WJY9 H0Z6G6
PDB
4N4A
E-value=4.46595e-09,
Score=148
Ontologies
GO
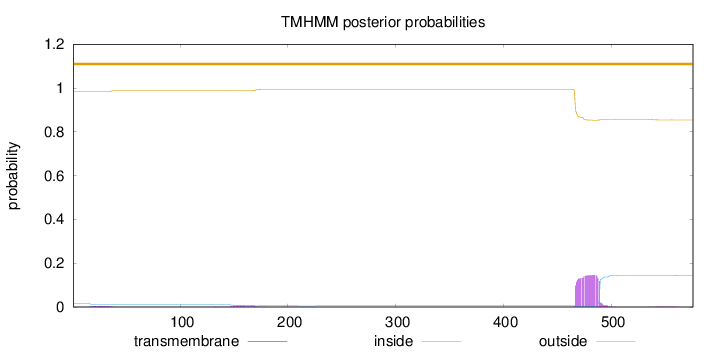
Topology
Subcellular location
Nucleus
Length:
576
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.53621
Exp number, first 60 AAs:
0.07994
Total prob of N-in:
0.01607
outside
1 - 576
Population Genetic Test Statistics
Pi
210.416914
Theta
199.599911
Tajima's D
0.157973
CLR
108.811759
CSRT
0.420178991050447
Interpretation
Uncertain
