Gene
KWMTBOMO08610
Pre Gene Modal
BGIBMGA007963
Annotation
PREDICTED:_raf_kinase?_effector_of_Ras_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.045
Sequence
CDS
ATGGGCTGCGTGTCGAAGCCGTCGCTGGCCATCGTGACGCAGTGGTGCGAGGGCTCGTCGCTCTACCAGCACCTGCACGTGCTCGAGACGCCCTTCCCCATCATATACCTCATCGACGTGGCCAGGCAGACGGCCCAGGGCATGGACTATCTGCACGCCAAGAACATCATACACAGAGATTTGAAATCAAATAACATATTTTTGAGAGATGACTGGTCGGTAAAGATTGGAGACTTTGGACTTGCTACTGCTAAAGTTCGGTGGTCAGAGTCGTCGTCGTCAGGCGGCGTGCAGTGGCAGCAGCCCACGGGCAGCATCCTGTGGATGGCGCCCGAGGTGATCCGCATGGAGGAGCCGGCGCCCTACACGTTCCACTCGGACGTGTACTCGTACGGCGTGGTGCTGTACGAGCTCATGGCGGGCGAGCTGCCCTACTCGCACCTCAACAACAAGGACCAGATCCTGTGGCTGGTGGGGCGCGGCCGCCTGCGCCCCGACGTGCGGCGCCTGCGCCCCGACGCGCCCGCGCAGCTCAAGCGCACCTTCTCCGAGTGCGTCGCCTTCGACCGCGCCGCGCGACCGCTCTTCCGCCAGGTGCTGGCGGCGCTGGAGGCGGTGCTGCGGCTGGTGCCCAAGATCACGCGCTCGGCGTCGGAGCCCGCGCTGTCGCACCCGCACCCGCGCGCCGCGCTCGACTACCTCGCCTACTCCTGCGCCTCGCCCAAGACGCCGGTCAACTTCCACTTCAACGCCGACACCAGCTTCCCCGCCTTCTACGGAATCCCAGTACCCAATCAGAGGCACCCCTAG
Protein
MGCVSKPSLAIVTQWCEGSSLYQHLHVLETPFPIIYLIDVARQTAQGMDYLHAKNIIHRDLKSNNIFLRDDWSVKIGDFGLATAKVRWSESSSSGGVQWQQPTGSILWMAPEVIRMEEPAPYTFHSDVYSYGVVLYELMAGELPYSHLNNKDQILWLVGRGRLRPDVRRLRPDAPAQLKRTFSECVAFDRAARPLFRQVLAALEAVLRLVPKITRSASEPALSHPHPRAALDYLAYSCASPKTPVNFHFNADTSFPAFYGIPVPNQRHP
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
E9JEI8
A0A2H1V261
A0A2A4JB31
A0A194PPH7
A0A0N0PES0
A0A212F6A7
+ More
A0A026WP12 A0A0L7R5I4 A0A310SGR0 A0A088ACF7 A0A0M9AAS9 A0A2A3EFS2 A0A154PI51 E1ZXD4 A0A3L8E3V7 K7ITS2 A0A1V9XPX4 A0A0J7NMM0 A0A2M4AVD8 A0A1W4WXJ6 A0A067QRW6 Q1DH71 A0A2M3ZIU7 A0A0P6DN88 A0A0C9RME2 Q7Q2L5 A0A1S4G0Q3 A0A182VJ19 A0A182TPA0 A0A182ML59 A0A1S4GMK6 A0A182LD89 A0A182HYC5 A0A0V0GAM4 A0A0P5L9P0 A0A182H0T5 T1HJK8 A0A084W1A1 A0A0M4M1V7 A0A023F485 A0A069DZF9 A0A182QWW8 A0A182IN34 V5GU10 W5JTG9 A0A3R7SUJ5 A0A2M4BFX8 A0A0A9X2K4 A0A1L8DL42 A0A336L0X1 A0A0P5NU23 A0A1D2MJ60 A0A131XP02 A0A0N8E8N3 A0A1Y1M7Y0 A0A1I8M2E9 E0VW01 A0A1B0CNY2 A0A2J7PSA6 A0A1Q3F2R8 A0A1Q3F2X7 A0A1E1XC06 A0A1B6I9J8 D6X4G2 E9GAH4 A0A1I8P278 A0A3S3PK80 A0A0L0BU75 B0WBK1 A0A0P4XGL5 A0A1Z5L444 A0A293LGS8 A0A131Z1W7 A0A224Z7T0 A0A226DE91 A0A1E1XM47 A0A2R5LNS9 A0A0A1X8T6 A0A034WUF5 A0A0P6FFZ4 A0A0K8V1V4 W8BRG2 A0A2R7WGR4 A0A0P6FEQ3 A0A131XUX6 A0A0P6F0R5 A0A0P5PY22 A0A0P5QKU5 A0A0P5GRN5 A0A164SHG4 A0A0P6FMN4 A0A0P6AA74 A0A0P6GXY8 A0A0P5GWL3 A0A0P6G6Y6 A0A3B3X4L4 A0A0P6E7T2 A0A0P5XM39
A0A026WP12 A0A0L7R5I4 A0A310SGR0 A0A088ACF7 A0A0M9AAS9 A0A2A3EFS2 A0A154PI51 E1ZXD4 A0A3L8E3V7 K7ITS2 A0A1V9XPX4 A0A0J7NMM0 A0A2M4AVD8 A0A1W4WXJ6 A0A067QRW6 Q1DH71 A0A2M3ZIU7 A0A0P6DN88 A0A0C9RME2 Q7Q2L5 A0A1S4G0Q3 A0A182VJ19 A0A182TPA0 A0A182ML59 A0A1S4GMK6 A0A182LD89 A0A182HYC5 A0A0V0GAM4 A0A0P5L9P0 A0A182H0T5 T1HJK8 A0A084W1A1 A0A0M4M1V7 A0A023F485 A0A069DZF9 A0A182QWW8 A0A182IN34 V5GU10 W5JTG9 A0A3R7SUJ5 A0A2M4BFX8 A0A0A9X2K4 A0A1L8DL42 A0A336L0X1 A0A0P5NU23 A0A1D2MJ60 A0A131XP02 A0A0N8E8N3 A0A1Y1M7Y0 A0A1I8M2E9 E0VW01 A0A1B0CNY2 A0A2J7PSA6 A0A1Q3F2R8 A0A1Q3F2X7 A0A1E1XC06 A0A1B6I9J8 D6X4G2 E9GAH4 A0A1I8P278 A0A3S3PK80 A0A0L0BU75 B0WBK1 A0A0P4XGL5 A0A1Z5L444 A0A293LGS8 A0A131Z1W7 A0A224Z7T0 A0A226DE91 A0A1E1XM47 A0A2R5LNS9 A0A0A1X8T6 A0A034WUF5 A0A0P6FFZ4 A0A0K8V1V4 W8BRG2 A0A2R7WGR4 A0A0P6FEQ3 A0A131XUX6 A0A0P6F0R5 A0A0P5PY22 A0A0P5QKU5 A0A0P5GRN5 A0A164SHG4 A0A0P6FMN4 A0A0P6AA74 A0A0P6GXY8 A0A0P5GWL3 A0A0P6G6Y6 A0A3B3X4L4 A0A0P6E7T2 A0A0P5XM39
Pubmed
19121390
21040523
26354079
22118469
24508170
20798317
+ More
30249741 20075255 28327890 24845553 17510324 12364791 20966253 26483478 24438588 25474469 26334808 25765539 20920257 23761445 25401762 26823975 27289101 28049606 28004739 25315136 20566863 28503490 18362917 19820115 21292972 26108605 28528879 26830274 28797301 29209593 25830018 25348373 24495485
30249741 20075255 28327890 24845553 17510324 12364791 20966253 26483478 24438588 25474469 26334808 25765539 20920257 23761445 25401762 26823975 27289101 28049606 28004739 25315136 20566863 28503490 18362917 19820115 21292972 26108605 28528879 26830274 28797301 29209593 25830018 25348373 24495485
EMBL
BABH01003305
GQ426305
ADM32531.1
ODYU01000345
SOQ34938.1
NWSH01002297
+ More
PCG68603.1 KQ459596 KPI95346.1 KQ459717 KPJ20286.1 AGBW02010064 OWR49271.1 KK107154 EZA56844.1 KQ414654 KOC66021.1 KQ760132 OAD61803.1 KQ435712 KOX79359.1 KZ288256 PBC30605.1 KQ434923 KZC11492.1 GL435030 EFN74224.1 QOIP01000001 RLU27095.1 MNPL01006238 OQR75536.1 LBMM01003264 KMQ93735.1 GGFK01011410 MBW44731.1 KK853027 KDR12291.1 CH899794 EAT32508.1 GGFM01007617 MBW28368.1 GDIQ01074538 JAN20199.1 GBYB01009489 GBYB01009490 GBYB01009492 JAG79256.1 JAG79257.1 JAG79259.1 AAAB01008968 EAA13186.4 AXCM01001310 APCN01006919 GECL01001009 JAP05115.1 GDIQ01177898 JAK73827.1 JXUM01102040 KQ564695 KXJ71888.1 ACPB03007057 ATLV01019249 KE525266 KFB43995.1 KR075836 ALE20555.1 GBBI01002883 JAC15829.1 GBGD01000710 JAC88179.1 AXCN02001895 GANP01010583 JAB73885.1 ADMH02000191 ETN67416.1 QCYY01001737 ROT75760.1 GGFJ01002819 MBW51960.1 GBHO01032269 GDHC01008017 JAG11335.1 JAQ10612.1 GFDF01007024 JAV07060.1 UFQS01001210 UFQS01001692 UFQT01001210 UFQT01001692 SSX09808.1 SSX11882.1 SSX29531.1 GDIQ01144560 JAL07166.1 LJIJ01001078 ODM93078.1 GEFH01001280 JAP67301.1 GDIQ01051004 JAN43733.1 GEZM01037775 JAV81939.1 AAZO01005734 DS235816 EEB17557.1 AJWK01021148 NEVH01021937 PNF19209.1 GFDL01013204 JAV21841.1 GFDL01013197 JAV21848.1 GFAC01002405 JAT96783.1 GECU01024095 JAS83611.1 KQ971380 EEZ97554.2 GL732537 EFX83242.1 NCKU01000141 RWS16960.1 JRES01001343 KNC23538.1 DS231879 EDS42440.1 GDIP01251159 JAI72242.1 GFJQ02004915 JAW02055.1 GFWV01003213 GFWV01012594 MAA27943.1 GEDV01004436 JAP84121.1 GFPF01011697 MAA22843.1 LNIX01000020 OXA43892.1 GFAA01003119 JAU00316.1 GGLE01007035 MBY11161.1 GBXI01011957 GBXI01007137 JAD02335.1 JAD07155.1 GAKP01001127 JAC57825.1 GDIQ01049258 JAN45479.1 GDHF01019402 GDHF01017651 GDHF01009403 GDHF01008431 GDHF01005532 JAI32912.1 JAI34663.1 JAI42911.1 JAI43883.1 JAI46782.1 GAMC01002626 GAMC01002625 JAC03931.1 KK854799 PTY18844.1 GDIQ01051003 JAN43734.1 GEFM01005674 JAP70122.1 GDIQ01069397 JAN25340.1 GDIQ01132086 JAL19640.1 GDIQ01133575 JAL18151.1 GDIQ01236690 JAK15035.1 LRGB01002019 KZS09635.1 GDIQ01048105 JAN46632.1 GDIP01032642 JAM71073.1 GDIQ01027750 JAN66987.1 GDIQ01236689 JAK15036.1 GDIQ01048104 JAN46633.1 GDIQ01081037 JAN13700.1 GDIP01070085 JAM33630.1
PCG68603.1 KQ459596 KPI95346.1 KQ459717 KPJ20286.1 AGBW02010064 OWR49271.1 KK107154 EZA56844.1 KQ414654 KOC66021.1 KQ760132 OAD61803.1 KQ435712 KOX79359.1 KZ288256 PBC30605.1 KQ434923 KZC11492.1 GL435030 EFN74224.1 QOIP01000001 RLU27095.1 MNPL01006238 OQR75536.1 LBMM01003264 KMQ93735.1 GGFK01011410 MBW44731.1 KK853027 KDR12291.1 CH899794 EAT32508.1 GGFM01007617 MBW28368.1 GDIQ01074538 JAN20199.1 GBYB01009489 GBYB01009490 GBYB01009492 JAG79256.1 JAG79257.1 JAG79259.1 AAAB01008968 EAA13186.4 AXCM01001310 APCN01006919 GECL01001009 JAP05115.1 GDIQ01177898 JAK73827.1 JXUM01102040 KQ564695 KXJ71888.1 ACPB03007057 ATLV01019249 KE525266 KFB43995.1 KR075836 ALE20555.1 GBBI01002883 JAC15829.1 GBGD01000710 JAC88179.1 AXCN02001895 GANP01010583 JAB73885.1 ADMH02000191 ETN67416.1 QCYY01001737 ROT75760.1 GGFJ01002819 MBW51960.1 GBHO01032269 GDHC01008017 JAG11335.1 JAQ10612.1 GFDF01007024 JAV07060.1 UFQS01001210 UFQS01001692 UFQT01001210 UFQT01001692 SSX09808.1 SSX11882.1 SSX29531.1 GDIQ01144560 JAL07166.1 LJIJ01001078 ODM93078.1 GEFH01001280 JAP67301.1 GDIQ01051004 JAN43733.1 GEZM01037775 JAV81939.1 AAZO01005734 DS235816 EEB17557.1 AJWK01021148 NEVH01021937 PNF19209.1 GFDL01013204 JAV21841.1 GFDL01013197 JAV21848.1 GFAC01002405 JAT96783.1 GECU01024095 JAS83611.1 KQ971380 EEZ97554.2 GL732537 EFX83242.1 NCKU01000141 RWS16960.1 JRES01001343 KNC23538.1 DS231879 EDS42440.1 GDIP01251159 JAI72242.1 GFJQ02004915 JAW02055.1 GFWV01003213 GFWV01012594 MAA27943.1 GEDV01004436 JAP84121.1 GFPF01011697 MAA22843.1 LNIX01000020 OXA43892.1 GFAA01003119 JAU00316.1 GGLE01007035 MBY11161.1 GBXI01011957 GBXI01007137 JAD02335.1 JAD07155.1 GAKP01001127 JAC57825.1 GDIQ01049258 JAN45479.1 GDHF01019402 GDHF01017651 GDHF01009403 GDHF01008431 GDHF01005532 JAI32912.1 JAI34663.1 JAI42911.1 JAI43883.1 JAI46782.1 GAMC01002626 GAMC01002625 JAC03931.1 KK854799 PTY18844.1 GDIQ01051003 JAN43734.1 GEFM01005674 JAP70122.1 GDIQ01069397 JAN25340.1 GDIQ01132086 JAL19640.1 GDIQ01133575 JAL18151.1 GDIQ01236690 JAK15035.1 LRGB01002019 KZS09635.1 GDIQ01048105 JAN46632.1 GDIP01032642 JAM71073.1 GDIQ01027750 JAN66987.1 GDIQ01236689 JAK15036.1 GDIQ01048104 JAN46633.1 GDIQ01081037 JAN13700.1 GDIP01070085 JAM33630.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000053097
+ More
UP000053825 UP000005203 UP000053105 UP000242457 UP000076502 UP000000311 UP000279307 UP000002358 UP000192247 UP000036403 UP000192223 UP000027135 UP000008820 UP000007062 UP000075903 UP000075902 UP000075883 UP000075882 UP000075840 UP000069940 UP000249989 UP000015103 UP000030765 UP000075886 UP000075880 UP000000673 UP000283509 UP000094527 UP000095301 UP000009046 UP000092461 UP000235965 UP000007266 UP000000305 UP000095300 UP000285301 UP000037069 UP000002320 UP000198287 UP000076858 UP000261480
UP000053825 UP000005203 UP000053105 UP000242457 UP000076502 UP000000311 UP000279307 UP000002358 UP000192247 UP000036403 UP000192223 UP000027135 UP000008820 UP000007062 UP000075903 UP000075902 UP000075883 UP000075882 UP000075840 UP000069940 UP000249989 UP000015103 UP000030765 UP000075886 UP000075880 UP000000673 UP000283509 UP000094527 UP000095301 UP000009046 UP000092461 UP000235965 UP000007266 UP000000305 UP000095300 UP000285301 UP000037069 UP000002320 UP000198287 UP000076858 UP000261480
PRIDE
Interpro
CDD
ProteinModelPortal
E9JEI8
A0A2H1V261
A0A2A4JB31
A0A194PPH7
A0A0N0PES0
A0A212F6A7
+ More
A0A026WP12 A0A0L7R5I4 A0A310SGR0 A0A088ACF7 A0A0M9AAS9 A0A2A3EFS2 A0A154PI51 E1ZXD4 A0A3L8E3V7 K7ITS2 A0A1V9XPX4 A0A0J7NMM0 A0A2M4AVD8 A0A1W4WXJ6 A0A067QRW6 Q1DH71 A0A2M3ZIU7 A0A0P6DN88 A0A0C9RME2 Q7Q2L5 A0A1S4G0Q3 A0A182VJ19 A0A182TPA0 A0A182ML59 A0A1S4GMK6 A0A182LD89 A0A182HYC5 A0A0V0GAM4 A0A0P5L9P0 A0A182H0T5 T1HJK8 A0A084W1A1 A0A0M4M1V7 A0A023F485 A0A069DZF9 A0A182QWW8 A0A182IN34 V5GU10 W5JTG9 A0A3R7SUJ5 A0A2M4BFX8 A0A0A9X2K4 A0A1L8DL42 A0A336L0X1 A0A0P5NU23 A0A1D2MJ60 A0A131XP02 A0A0N8E8N3 A0A1Y1M7Y0 A0A1I8M2E9 E0VW01 A0A1B0CNY2 A0A2J7PSA6 A0A1Q3F2R8 A0A1Q3F2X7 A0A1E1XC06 A0A1B6I9J8 D6X4G2 E9GAH4 A0A1I8P278 A0A3S3PK80 A0A0L0BU75 B0WBK1 A0A0P4XGL5 A0A1Z5L444 A0A293LGS8 A0A131Z1W7 A0A224Z7T0 A0A226DE91 A0A1E1XM47 A0A2R5LNS9 A0A0A1X8T6 A0A034WUF5 A0A0P6FFZ4 A0A0K8V1V4 W8BRG2 A0A2R7WGR4 A0A0P6FEQ3 A0A131XUX6 A0A0P6F0R5 A0A0P5PY22 A0A0P5QKU5 A0A0P5GRN5 A0A164SHG4 A0A0P6FMN4 A0A0P6AA74 A0A0P6GXY8 A0A0P5GWL3 A0A0P6G6Y6 A0A3B3X4L4 A0A0P6E7T2 A0A0P5XM39
A0A026WP12 A0A0L7R5I4 A0A310SGR0 A0A088ACF7 A0A0M9AAS9 A0A2A3EFS2 A0A154PI51 E1ZXD4 A0A3L8E3V7 K7ITS2 A0A1V9XPX4 A0A0J7NMM0 A0A2M4AVD8 A0A1W4WXJ6 A0A067QRW6 Q1DH71 A0A2M3ZIU7 A0A0P6DN88 A0A0C9RME2 Q7Q2L5 A0A1S4G0Q3 A0A182VJ19 A0A182TPA0 A0A182ML59 A0A1S4GMK6 A0A182LD89 A0A182HYC5 A0A0V0GAM4 A0A0P5L9P0 A0A182H0T5 T1HJK8 A0A084W1A1 A0A0M4M1V7 A0A023F485 A0A069DZF9 A0A182QWW8 A0A182IN34 V5GU10 W5JTG9 A0A3R7SUJ5 A0A2M4BFX8 A0A0A9X2K4 A0A1L8DL42 A0A336L0X1 A0A0P5NU23 A0A1D2MJ60 A0A131XP02 A0A0N8E8N3 A0A1Y1M7Y0 A0A1I8M2E9 E0VW01 A0A1B0CNY2 A0A2J7PSA6 A0A1Q3F2R8 A0A1Q3F2X7 A0A1E1XC06 A0A1B6I9J8 D6X4G2 E9GAH4 A0A1I8P278 A0A3S3PK80 A0A0L0BU75 B0WBK1 A0A0P4XGL5 A0A1Z5L444 A0A293LGS8 A0A131Z1W7 A0A224Z7T0 A0A226DE91 A0A1E1XM47 A0A2R5LNS9 A0A0A1X8T6 A0A034WUF5 A0A0P6FFZ4 A0A0K8V1V4 W8BRG2 A0A2R7WGR4 A0A0P6FEQ3 A0A131XUX6 A0A0P6F0R5 A0A0P5PY22 A0A0P5QKU5 A0A0P5GRN5 A0A164SHG4 A0A0P6FMN4 A0A0P6AA74 A0A0P6GXY8 A0A0P5GWL3 A0A0P6G6Y6 A0A3B3X4L4 A0A0P6E7T2 A0A0P5XM39
Ontologies
GO
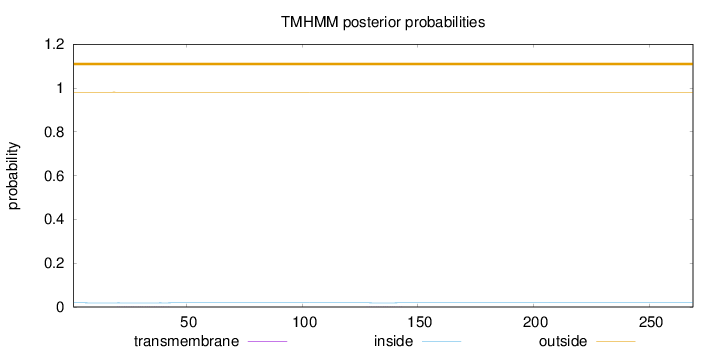
Topology
Length:
269
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0192
Exp number, first 60 AAs:
0.01509
Total prob of N-in:
0.01886
outside
1 - 269
Population Genetic Test Statistics
Pi
184.730571
Theta
163.238139
Tajima's D
0.892781
CLR
0.63117
CSRT
0.625368731563422
Interpretation
Uncertain
