Gene
KWMTBOMO08605
Pre Gene Modal
BGIBMGA007617
Annotation
PREDICTED:_nose_resistant_to_fluoxetine_protein_6-like_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.852
Sequence
CDS
ATGACTCCTTTAGTGTTTTTATTTGTATGTGTAGTTTTAGTTAATAGTGCCGAACTATCGAACGGGAGAGATAACGTCAGAGAAGAGAAGTTGAGAAGCGGCTTAATAGATGCCTTTCAGAATATCAAGAAAGCCTCCGCGAAAGGCGGGATTGATGATGAGAGTCCCCTGACCTCAGACCTCGTGGGGTCTATGATACAAGCCCCGAACACGAGCAGAGACGAGAAATCGACAACCGAAGACTCCTTCGACTTGTTCGCTGATGATGACGGTCTCTCCTTTGCTGTAAACAGCAAAATGGAAGATGAACAGAAGCAGCCCACCACGGAGAAAGCTCACAACGTTCAACATATTGTAAAAACGACAAAACTAGAGAAGACAACTTCGAAACCGAACAAAGACGTCTTCGGTAAATTAAACGACTCGATCAGGAGTGCGGTCAACGAGTCCAGGATGGCTCTGAAGAGGGACGGTTTGAACATGACTCAGTTTACGGGTATTGAGAGTATGCTGAGGGTGTTCGACGCGGAAATCCTCATGAGGCAGTGGAGAGATCTACAAGGTGCGCTCACGGGGGAGTGTAAGAAGGAGATGCAGATGTACGTCGATGGACTTCATGAAAAAAGACTATGGGCTTTGAAGATGGAAGATGCCGGTGGTCGCTACACCTCGATGTTCTACTGGGGGAACAACTACTGGACGGGATCCGCTGAGCTCTGCCAGGTCCTCAACCAACAGCAGGCGACTCCAGCACCTTCTAATAAAAGTTCCAGTTCCCAGATCTTCAGCGAGTGGCGTCAGGATGTGTCCATGACCGGGTCGGGACCGCCTTTCCGGGCAGCTTTCTACACAGTCCGAGTTTCGGTCACCTCAGAACTACCGGAAATTGTCAAAACTCGCCGTACTCTCCACCTTGGTCTCTGTCTCCCGTACTCCTGCTCTGGCGAGGCTGTCTCCGCGTTAGTCGGTAAAGCGAAGGCCCCTCTACTCCAACACTCCGTTCTCGCCGTCCGCACTCCATTCCACGGAGGATACTCTTACATCGAAGATCCTACTTTCCAAACACTTCTCGGCGTCTGCTGTATTCTAGCAGTTGTACTGATAACAGCCACCGTCTACGACGTGAGGATATCTCAAGAGATTGGACGTCGGAAGCAGCGAGCAGCCAATATCGCGACCAACACACCATCCGACCTCAAGCTGGACCTCAACGCGCTTGAAGATATCGCCGTTGCCAACGGTAAATCTATTTACAACGTCAATAACAACATCCCTATCGGCAACTCGTCCAAGGAACAGCTCACGGATACAGCCTCCACCTCCACAGAGGACGAGCTCAAGCTCAGCATATGGTCGAAGCTCTTGCTGTCGTTCTCCATGAGGACGAACATAATTCAGATCTTCGACCAGAGCGTGGGCGCCGACACCGTGCCGGTTGTGCACGGACTCAGAACCCTCTCCATGGTCTGGGTGATATTTGGACACACTTGCATCGTTGTATTCAAGTACGCAGACAACACAGCTTTAAGAGCTATTCTAGAGAAAAGCTTCTGGTTCCAATTGATCATAAGCGCGGTCTATAGCGTTGACACATTCTTCTGTTTGGGTGGAATGCTGGTTACATTTCTGTATTTCCGATCGAACGCTAAAGGCAAGTTGGATCGGCTGACGAAGGGCAGACGGAACTTCGCCTCGGGAGCGCTGCAGTTCTTAGGACTTCTTGGCTACAGATTTGCCAGGCTCACGGCCCCCTACCTGTTTGTCCTGGGCGTGGTGGAGGTCACCATGAAATGGTTCGCACACAACGCTGTCTTCGAACCCCCTGCGATGGATTACGAGACCTGCCCCAAGTACTGGTGGAGGAATCTGCTCTACATCAACACTTTGTTCCCGGTCGAGCAGATGTGCATGTTGTGGAGTTGGTATCTATCGGACGACACCCAGTTCTACGCTGTCAGCGCCATCCTTCTCATCTTATCGACGAGTCATTTCAAACTGTCAGCGTCGTTAACTGGCGTATTTTTCGTGTCGTCCCTGGTGACCACGGGCTACGTGTCCTGGAGCAACGACCACATCCCGAACAGCGAGGATCCCTTCACACAGTTCGACAAGATCTACGACAAGCCCTGGACTAGACTTGGCCCCTATTTGGTCGGCATGCTCACCGGTTGGATCCTCTTCAAGACTAACTTGAAGATTCGAATGAGCAGGATATGGGCGTGGGTTGGGTGGTTGATATGCGCCGCTACGTTTTTGTTCCTGATCTTTGGGCTCTACAACACAGAGCTGAGCGTGGCGTCAGCTGCGGTGTACAGCGCGCTCTCCCACTCTCTGTGGGCTGCCTGCGTGGGCTGGGTCATCATTGCATGCTCCACTGGACATGGAGGCTGGGTGCGGCCGCTGCTGGCGTCGCCCGTGCTGTACCCGTTCTCGCGCGTGACGTACTGCGCCTACCTCGTGCACCCCGTCGTGCTGCGCTACGTCGCGATGCACCTCACGCACCCCATCCACCTGGGAGAGCTGCTCGTGTTCGTCCTTTTCCTGGGACTAACGGTCATCTCATACTTCCTGGCGTTCCTGGTCTCTGTAGCCTTCGAAGCTCCCATCGTCATGATGCTTAAACTCTTATCGCCACAGAAGAAGTCTCACTGA
Protein
MTPLVFLFVCVVLVNSAELSNGRDNVREEKLRSGLIDAFQNIKKASAKGGIDDESPLTSDLVGSMIQAPNTSRDEKSTTEDSFDLFADDDGLSFAVNSKMEDEQKQPTTEKAHNVQHIVKTTKLEKTTSKPNKDVFGKLNDSIRSAVNESRMALKRDGLNMTQFTGIESMLRVFDAEILMRQWRDLQGALTGECKKEMQMYVDGLHEKRLWALKMEDAGGRYTSMFYWGNNYWTGSAELCQVLNQQQATPAPSNKSSSSQIFSEWRQDVSMTGSGPPFRAAFYTVRVSVTSELPEIVKTRRTLHLGLCLPYSCSGEAVSALVGKAKAPLLQHSVLAVRTPFHGGYSYIEDPTFQTLLGVCCILAVVLITATVYDVRISQEIGRRKQRAANIATNTPSDLKLDLNALEDIAVANGKSIYNVNNNIPIGNSSKEQLTDTASTSTEDELKLSIWSKLLLSFSMRTNIIQIFDQSVGADTVPVVHGLRTLSMVWVIFGHTCIVVFKYADNTALRAILEKSFWFQLIISAVYSVDTFFCLGGMLVTFLYFRSNAKGKLDRLTKGRRNFASGALQFLGLLGYRFARLTAPYLFVLGVVEVTMKWFAHNAVFEPPAMDYETCPKYWWRNLLYINTLFPVEQMCMLWSWYLSDDTQFYAVSAILLILSTSHFKLSASLTGVFFVSSLVTTGYVSWSNDHIPNSEDPFTQFDKIYDKPWTRLGPYLVGMLTGWILFKTNLKIRMSRIWAWVGWLICAATFLFLIFGLYNTELSVASAAVYSALSHSLWAACVGWVIIACSTGHGGWVRPLLASPVLYPFSRVTYCAYLVHPVVLRYVAMHLTHPIHLGELLVFVLFLGLTVISYFLAFLVSVAFEAPIVMMLKLLSPQKKSH
Summary
Uniprot
H9JDM0
A0A2A4JB09
A0A194PPV0
A0A212F6A9
A0A0N0PEP8
A0A232EUB1
+ More
A0A182R0W3 A0A182YB96 E2B9T3 A0A182L9B9 A0A182VJM0 A0A182HL05 D2A530 K7ITS0 A0A182FRS1 A0A1W7R5K7 A0A3L8E4G7 Q7PXM9 A0A182J9S8 A0A026WLC5 E9J424 A0A1I8MI13 A0A158NQM6 A0A195BQZ0 A0A0L7R5C7 A0A1I8NS18 W8B935 A0A0K8V9B3 A0A195DI53 A0A195ERS9 A0A195D3P0 B5DU10 A0A151XB20 A0A0A1WIC4 Q172T8 A0A2J7QRL5 A0A0L0CDB9 A0A1B6EC37 A0A182ME42 B4NPE5 A0A154PI16 A0A1B6LFZ8 A0A1J1J0V4 A0A182NA19 A0A088ACB9 A0A182TKQ5 A0A2A3EFW4 A0A182PH02 W5JMI9 A0A336M2H7 A0A0J7KR15 A0A034VCF1 A0A1A9W9H0 A0A182R512 A0A336KG07 A0A1B0FQH3 A0A1S4FG92 A0A0N0BJM3 F4WKF7 J9KAG5 A0A182K452 E2AN08 A0A2H8TDX4 A0A310SV83 A0A182XA26 A0A182VWD5 A0A1B0ANF3 A0A1A9VRM5 A0A1A9XGZ2 A0A0A9XFD3 A0A2H8TX63 J9JXD2 A0A2S2N858 A0A026W0K9
A0A182R0W3 A0A182YB96 E2B9T3 A0A182L9B9 A0A182VJM0 A0A182HL05 D2A530 K7ITS0 A0A182FRS1 A0A1W7R5K7 A0A3L8E4G7 Q7PXM9 A0A182J9S8 A0A026WLC5 E9J424 A0A1I8MI13 A0A158NQM6 A0A195BQZ0 A0A0L7R5C7 A0A1I8NS18 W8B935 A0A0K8V9B3 A0A195DI53 A0A195ERS9 A0A195D3P0 B5DU10 A0A151XB20 A0A0A1WIC4 Q172T8 A0A2J7QRL5 A0A0L0CDB9 A0A1B6EC37 A0A182ME42 B4NPE5 A0A154PI16 A0A1B6LFZ8 A0A1J1J0V4 A0A182NA19 A0A088ACB9 A0A182TKQ5 A0A2A3EFW4 A0A182PH02 W5JMI9 A0A336M2H7 A0A0J7KR15 A0A034VCF1 A0A1A9W9H0 A0A182R512 A0A336KG07 A0A1B0FQH3 A0A1S4FG92 A0A0N0BJM3 F4WKF7 J9KAG5 A0A182K452 E2AN08 A0A2H8TDX4 A0A310SV83 A0A182XA26 A0A182VWD5 A0A1B0ANF3 A0A1A9VRM5 A0A1A9XGZ2 A0A0A9XFD3 A0A2H8TX63 J9JXD2 A0A2S2N858 A0A026W0K9
Pubmed
EMBL
BABH01003309
NWSH01002297
PCG68610.1
KQ459596
KPI95342.1
AGBW02010064
+ More
OWR49275.1 KQ459717 KPJ20289.1 NNAY01002149 OXU21931.1 AXCN02001522 GL446605 EFN87520.1 APCN01000876 KQ971361 EFA05306.2 GEHC01001350 JAV46295.1 QOIP01000001 RLU27557.1 AAAB01008987 EAA01818.5 KK107154 EZA56840.1 GL768069 EFZ12494.1 ADTU01023412 ADTU01023413 ADTU01023414 KQ976419 KYM89271.1 KQ414654 KOC66024.1 GAMC01012972 JAB93583.1 GDHF01016867 JAI35447.1 KQ980824 KYN12521.1 KQ982021 KYN30584.1 KQ976885 KYN07512.1 CH476200 EDY71464.1 KQ982335 KYQ57544.1 GBXI01016037 JAC98254.1 CH477432 EAT41009.1 NEVH01011893 PNF31216.1 JRES01000557 KNC30260.1 GEDC01001793 JAS35505.1 AXCM01012822 CH964291 EDW86385.1 KQ434923 KZC11495.1 GEBQ01017372 JAT22605.1 CVRI01000063 CRL04481.1 KZ288256 PBC30608.1 ADMH02001137 ETN63969.1 UFQS01000320 UFQT01000320 SSX02841.1 SSX23209.1 LBMM01004227 KMQ92679.1 GAKP01019754 GAKP01019753 JAC39198.1 SSX02842.1 SSX23210.1 CCAG010010824 KQ435712 KOX79362.1 GL888199 EGI65295.1 ABLF02036237 GL441034 EFN65166.1 GFXV01000502 MBW12307.1 KQ760132 OAD61806.1 JXJN01000809 GBHO01026096 JAG17508.1 GFXV01006695 MBW18500.1 ABLF02038822 GGMR01000701 MBY13320.1 KK107539 QOIP01000006 EZA49101.1 RLU21565.1
OWR49275.1 KQ459717 KPJ20289.1 NNAY01002149 OXU21931.1 AXCN02001522 GL446605 EFN87520.1 APCN01000876 KQ971361 EFA05306.2 GEHC01001350 JAV46295.1 QOIP01000001 RLU27557.1 AAAB01008987 EAA01818.5 KK107154 EZA56840.1 GL768069 EFZ12494.1 ADTU01023412 ADTU01023413 ADTU01023414 KQ976419 KYM89271.1 KQ414654 KOC66024.1 GAMC01012972 JAB93583.1 GDHF01016867 JAI35447.1 KQ980824 KYN12521.1 KQ982021 KYN30584.1 KQ976885 KYN07512.1 CH476200 EDY71464.1 KQ982335 KYQ57544.1 GBXI01016037 JAC98254.1 CH477432 EAT41009.1 NEVH01011893 PNF31216.1 JRES01000557 KNC30260.1 GEDC01001793 JAS35505.1 AXCM01012822 CH964291 EDW86385.1 KQ434923 KZC11495.1 GEBQ01017372 JAT22605.1 CVRI01000063 CRL04481.1 KZ288256 PBC30608.1 ADMH02001137 ETN63969.1 UFQS01000320 UFQT01000320 SSX02841.1 SSX23209.1 LBMM01004227 KMQ92679.1 GAKP01019754 GAKP01019753 JAC39198.1 SSX02842.1 SSX23210.1 CCAG010010824 KQ435712 KOX79362.1 GL888199 EGI65295.1 ABLF02036237 GL441034 EFN65166.1 GFXV01000502 MBW12307.1 KQ760132 OAD61806.1 JXJN01000809 GBHO01026096 JAG17508.1 GFXV01006695 MBW18500.1 ABLF02038822 GGMR01000701 MBY13320.1 KK107539 QOIP01000006 EZA49101.1 RLU21565.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000215335
+ More
UP000075886 UP000076408 UP000008237 UP000075882 UP000075903 UP000075840 UP000007266 UP000002358 UP000069272 UP000279307 UP000007062 UP000075880 UP000053097 UP000095301 UP000005205 UP000078540 UP000053825 UP000095300 UP000078492 UP000078541 UP000078542 UP000001819 UP000075809 UP000008820 UP000235965 UP000037069 UP000075883 UP000007798 UP000076502 UP000183832 UP000075884 UP000005203 UP000075902 UP000242457 UP000075885 UP000000673 UP000036403 UP000091820 UP000075900 UP000092444 UP000053105 UP000007755 UP000007819 UP000075881 UP000000311 UP000076407 UP000075920 UP000092460 UP000078200 UP000092443
UP000075886 UP000076408 UP000008237 UP000075882 UP000075903 UP000075840 UP000007266 UP000002358 UP000069272 UP000279307 UP000007062 UP000075880 UP000053097 UP000095301 UP000005205 UP000078540 UP000053825 UP000095300 UP000078492 UP000078541 UP000078542 UP000001819 UP000075809 UP000008820 UP000235965 UP000037069 UP000075883 UP000007798 UP000076502 UP000183832 UP000075884 UP000005203 UP000075902 UP000242457 UP000075885 UP000000673 UP000036403 UP000091820 UP000075900 UP000092444 UP000053105 UP000007755 UP000007819 UP000075881 UP000000311 UP000076407 UP000075920 UP000092460 UP000078200 UP000092443
PRIDE
Pfam
PF01757 Acyl_transf_3
ProteinModelPortal
H9JDM0
A0A2A4JB09
A0A194PPV0
A0A212F6A9
A0A0N0PEP8
A0A232EUB1
+ More
A0A182R0W3 A0A182YB96 E2B9T3 A0A182L9B9 A0A182VJM0 A0A182HL05 D2A530 K7ITS0 A0A182FRS1 A0A1W7R5K7 A0A3L8E4G7 Q7PXM9 A0A182J9S8 A0A026WLC5 E9J424 A0A1I8MI13 A0A158NQM6 A0A195BQZ0 A0A0L7R5C7 A0A1I8NS18 W8B935 A0A0K8V9B3 A0A195DI53 A0A195ERS9 A0A195D3P0 B5DU10 A0A151XB20 A0A0A1WIC4 Q172T8 A0A2J7QRL5 A0A0L0CDB9 A0A1B6EC37 A0A182ME42 B4NPE5 A0A154PI16 A0A1B6LFZ8 A0A1J1J0V4 A0A182NA19 A0A088ACB9 A0A182TKQ5 A0A2A3EFW4 A0A182PH02 W5JMI9 A0A336M2H7 A0A0J7KR15 A0A034VCF1 A0A1A9W9H0 A0A182R512 A0A336KG07 A0A1B0FQH3 A0A1S4FG92 A0A0N0BJM3 F4WKF7 J9KAG5 A0A182K452 E2AN08 A0A2H8TDX4 A0A310SV83 A0A182XA26 A0A182VWD5 A0A1B0ANF3 A0A1A9VRM5 A0A1A9XGZ2 A0A0A9XFD3 A0A2H8TX63 J9JXD2 A0A2S2N858 A0A026W0K9
A0A182R0W3 A0A182YB96 E2B9T3 A0A182L9B9 A0A182VJM0 A0A182HL05 D2A530 K7ITS0 A0A182FRS1 A0A1W7R5K7 A0A3L8E4G7 Q7PXM9 A0A182J9S8 A0A026WLC5 E9J424 A0A1I8MI13 A0A158NQM6 A0A195BQZ0 A0A0L7R5C7 A0A1I8NS18 W8B935 A0A0K8V9B3 A0A195DI53 A0A195ERS9 A0A195D3P0 B5DU10 A0A151XB20 A0A0A1WIC4 Q172T8 A0A2J7QRL5 A0A0L0CDB9 A0A1B6EC37 A0A182ME42 B4NPE5 A0A154PI16 A0A1B6LFZ8 A0A1J1J0V4 A0A182NA19 A0A088ACB9 A0A182TKQ5 A0A2A3EFW4 A0A182PH02 W5JMI9 A0A336M2H7 A0A0J7KR15 A0A034VCF1 A0A1A9W9H0 A0A182R512 A0A336KG07 A0A1B0FQH3 A0A1S4FG92 A0A0N0BJM3 F4WKF7 J9KAG5 A0A182K452 E2AN08 A0A2H8TDX4 A0A310SV83 A0A182XA26 A0A182VWD5 A0A1B0ANF3 A0A1A9VRM5 A0A1A9XGZ2 A0A0A9XFD3 A0A2H8TX63 J9JXD2 A0A2S2N858 A0A026W0K9
Ontologies
GO
Topology
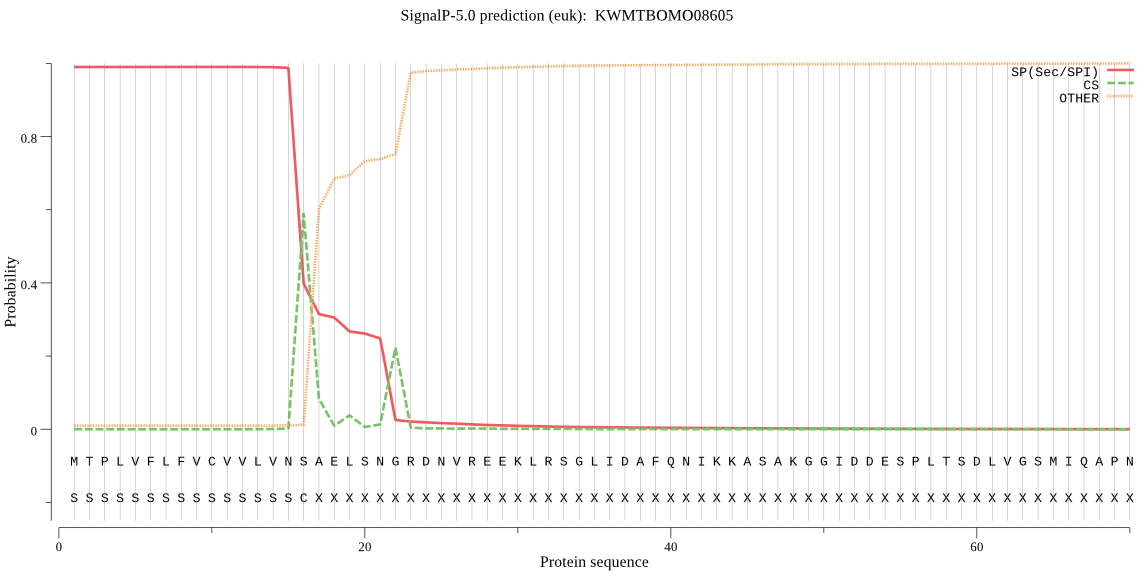
SignalP
Position: 1 - 16,
Likelihood: 0.989726
Length:
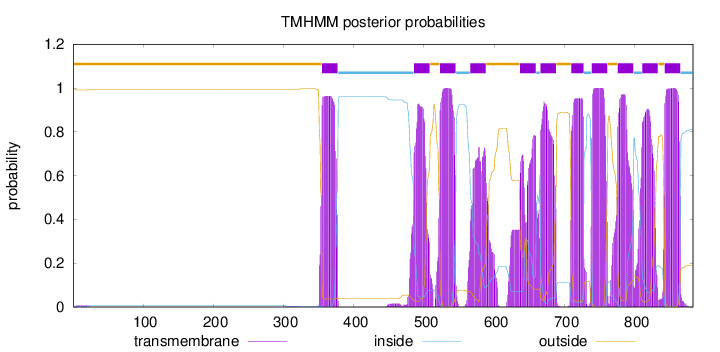
883
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
232.0798
Exp number, first 60 AAs:
0.12728
Total prob of N-in:
0.00443
outside
1 - 354
TMhelix
355 - 377
inside
378 - 485
TMhelix
486 - 508
outside
509 - 522
TMhelix
523 - 545
inside
546 - 565
TMhelix
566 - 588
outside
589 - 636
TMhelix
637 - 659
inside
660 - 665
TMhelix
666 - 688
outside
689 - 709
TMhelix
710 - 727
inside
728 - 738
TMhelix
739 - 761
outside
762 - 775
TMhelix
776 - 798
inside
799 - 810
TMhelix
811 - 833
outside
834 - 842
TMhelix
843 - 865
inside
866 - 883
Population Genetic Test Statistics
Pi
228.176086
Theta
152.748581
Tajima's D
0.194274
CLR
0.45764
CSRT
0.420428978551072
Interpretation
Uncertain
