Gene
KWMTBOMO08604
Pre Gene Modal
BGIBMGA007967
Annotation
PREDICTED:_dual_specificity_protein_phosphatase_12-like_[Papilio_machaon]
Full name
Dual specificity protein phosphatase MPK-4
Location in the cell
Cytoplasmic Reliability : 1.257 Nuclear Reliability : 1.734
Sequence
CDS
ATGGCGAAACAAGTTGCTGAAGTAGAAATACATGCTTCAGACTCCCAGGAGACCCTTGTGGACGACAATCTGGAGGTCAGCGTGGATCTGATTGACAAAAATCTTTATTTAGGAAACCTGGCATGTGCACACAGTCAAGCGACAATACAAACGCTTGGAATAACTCACATCTTGACAGTGGACTTGGTTCCTTTACCGAGGATTATACTGGAACGTCCTCATCTCATATTTAGATATGTGAAATTGGCTGATGTGCCCAAAGAAGACCTAATAAGTCATCTTCCCACTACGAACAATTATATAAGTGAAGCCATCGACAGTGGAGGAACTATATTAGTGCATTGTTATTTTGGTGTATCCCGTTCAGCAGCTGTTGTTATAGCATACATAATGAAGAAGTACGGGCTGTGTTACGAGGAAGCCTTCTCGCTGGTCAAAAACAAGAGACGGTTCATAGGTCCAAATGTAGGATTTGTGGCACAGCTCAAATTGTACGGGCACATGGGATGTGCGATTAACAAAGATGACCCTAGATACAAACAGTTTAGGTTGAAAATGGCGGGCCAAAAGTTAAAACAAGTGAAGATCCTGCCGCAAGTGTTCTGGGAGCTGGTGAAGCCGGACCCGGGCGTGGTCCGCGAGCGCCCCTCGCCGCACGTGTACCGCTGCGTGCGCTGCCGCCGCGTGCTCGCCGCGCACAACAACCTCATCCCCCACCTGCCGCGCCCGCTCAAGCTGCAACTCGCCAAGAAAGGTATAAGGCCCCCTCCGAGTAAACTGACGGGTTTGACCTGCGTTGAAAACGGCGAGATCCTGATACAGAAGTTGAAGAGCCTCGCCTGTCAGATCCTCGAGAGCGACGAGCAGGCTGACGGGGGACACGGCGGCGACGAAATGGAAGGAAGCAATCAGGAGTTACTGGACGGTTACAACGAGCAGAACCTGGTGGACGGCAGTGTAGTGCCGCGGGAATGCGAGACGATATGCCGACAGATGTGGTTCGTGGAGCCGATGGCGTGGATGAGTGACGTCACGCACCAGCCCAGCGGGAAGCTGCACTGTCCCAATTGCAAGAACAAGATCGGGAGCTTTAGTTGGGTTATGGGTTGCAAATGTCCGTGCGGTCAGAAAGTGGCTCCAGCTTTCTATTTAGTACCATCCAAAGTGGAGAGATCTAATATAGTTCAAAACGTACAAACCACTGTATAG
Protein
MAKQVAEVEIHASDSQETLVDDNLEVSVDLIDKNLYLGNLACAHSQATIQTLGITHILTVDLVPLPRIILERPHLIFRYVKLADVPKEDLISHLPTTNNYISEAIDSGGTILVHCYFGVSRSAAVVIAYIMKKYGLCYEEAFSLVKNKRRFIGPNVGFVAQLKLYGHMGCAINKDDPRYKQFRLKMAGQKLKQVKILPQVFWELVKPDPGVVRERPSPHVYRCVRCRRVLAAHNNLIPHLPRPLKLQLAKKGIRPPPSKLTGLTCVENGEILIQKLKSLACQILESDEQADGGHGGDEMEGSNQELLDGYNEQNLVDGSVVPRECETICRQMWFVEPMAWMSDVTHQPSGKLHCPNCKNKIGSFSWVMGCKCPCGQKVAPAFYLVPSKVERSNIVQNVQTTV
Summary
Description
Dual specificity phosphatase; can dephosphorylate both phosphotyrosine and phosphoserine or phosphothreonine residues. May suppress bsk/JNK activation during the immune response.
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
Subunit
Interacts (via tyrosine-protein phosphatase domain) with bsk/JNK; the interaction dephosphorylates bsk.
Similarity
Belongs to the protein-tyrosine phosphatase family. Non-receptor class dual specificity subfamily.
Keywords
Complete proteome
Cytoplasm
Hydrolase
Nucleus
Protein phosphatase
Reference proteome
Feature
chain Dual specificity protein phosphatase MPK-4
Uniprot
H9JEM0
A0A0N1IIT2
A0A194PW30
A0A2A4J9H9
A0A2H1WMR1
A0A212F6B2
+ More
A0A212F5D4 A0A0K8WHM0 A0A034W6R8 A0A437BDN4 B4NBU5 A0A0A1XRF2 A0A1I8Q332 A0A1W4VTG2 A0A1I8MD03 A0A0L0BVQ5 A0A1A9ZKU1 A0A1B0G8N9 A0A1B0BDL7 A0A1A9YMH8 B3MQB2 A0A0M5J635 E9IMS5 Q9VWF4 W8BQB5 B4I785 Q29IU9 B4PZF3 B3NVR0 A0A310SQ12 W5JB05 A0A0K8TNP7 F4WSV4 A0A2A3EGY1 A0A151J252 A0A195AZ10 A0A195D197 A0A088AW72 A0A158NH25 A0A182PNB8 A0A182XGH0 A0A1W5CAG9 A0A182QTS2 B4JM13 A0A182HZV6 A0A182M4P1 A0A195F5L2 B4MAI9 A0A1A9WVL4 A0A2M4BQ41 A0A182K5T2 A0A182JAV3 A0A182F7Z0 A0A182LQY6 A0A1S4GEJ6 A0A151XDD5 E2B2B0 A0A182N6A3 B4L676 A0A084W9U4 A0A182USS8 A0A182RT75 A0A1J1IGV1 Q7PYU3 A0A026W1G7 A0A182SKL9 A0A1A9UW41 Q16JJ4 A0A182XW25 A0A182G1Q7 E2AJC7 A0A182VRE3 A0A0K8UFZ6 A0A1Q3EWD7 B0XGR0 A0A0C9PJ41 A0A0L0CEX1 A0A226EX64 A0A3B0L0B5
A0A212F5D4 A0A0K8WHM0 A0A034W6R8 A0A437BDN4 B4NBU5 A0A0A1XRF2 A0A1I8Q332 A0A1W4VTG2 A0A1I8MD03 A0A0L0BVQ5 A0A1A9ZKU1 A0A1B0G8N9 A0A1B0BDL7 A0A1A9YMH8 B3MQB2 A0A0M5J635 E9IMS5 Q9VWF4 W8BQB5 B4I785 Q29IU9 B4PZF3 B3NVR0 A0A310SQ12 W5JB05 A0A0K8TNP7 F4WSV4 A0A2A3EGY1 A0A151J252 A0A195AZ10 A0A195D197 A0A088AW72 A0A158NH25 A0A182PNB8 A0A182XGH0 A0A1W5CAG9 A0A182QTS2 B4JM13 A0A182HZV6 A0A182M4P1 A0A195F5L2 B4MAI9 A0A1A9WVL4 A0A2M4BQ41 A0A182K5T2 A0A182JAV3 A0A182F7Z0 A0A182LQY6 A0A1S4GEJ6 A0A151XDD5 E2B2B0 A0A182N6A3 B4L676 A0A084W9U4 A0A182USS8 A0A182RT75 A0A1J1IGV1 Q7PYU3 A0A026W1G7 A0A182SKL9 A0A1A9UW41 Q16JJ4 A0A182XW25 A0A182G1Q7 E2AJC7 A0A182VRE3 A0A0K8UFZ6 A0A1Q3EWD7 B0XGR0 A0A0C9PJ41 A0A0L0CEX1 A0A226EX64 A0A3B0L0B5
EC Number
3.1.3.16
3.1.3.48
3.1.3.48
Pubmed
EMBL
BABH01003312
BABH01003313
KQ459717
KPJ20291.1
KQ459596
KPI95340.1
+ More
NWSH01002297 PCG68611.1 ODYU01009703 SOQ54318.1 AGBW02010064 OWR49276.1 AGBW02010203 OWR48940.1 GDHF01001663 JAI50651.1 GAKP01009122 JAC49830.1 RSAL01000083 RVE48458.1 CH964239 EDW82304.1 GBXI01012330 GBXI01000453 JAD01962.1 JAD13839.1 JRES01001264 KNC24101.1 CCAG010000297 JXJN01012511 CH902621 EDV44538.1 CP012528 ALC49024.1 GL764255 EFZ18155.1 AE014298 BT031134 AY059453 AAL13359.1 ABX00756.1 GAMC01007563 GAMC01007562 JAB98993.1 CH480823 EDW56183.1 CH379063 EAL32554.2 CM000162 EDX02109.1 CH954180 EDV46725.1 KQ761783 OAD56995.1 ADMH02002023 ETN59944.1 GDAI01001857 JAI15746.1 GL888327 EGI62761.1 KZ288256 PBC30542.1 KQ980447 KYN16030.1 KQ976698 KYM77441.1 KQ976973 KYN06678.1 ADTU01015494 AAAB01008987 EGK97380.1 AXCN02000278 CH916371 EDV91774.1 APCN01002014 AXCM01000395 KQ981768 KYN35880.1 CH940655 EDW66248.1 GGFJ01005900 MBW55041.1 KQ982294 KYQ58359.1 GL445130 EFN90147.1 CH933812 EDW05872.1 ATLV01021913 KE525325 KFB46988.1 CVRI01000050 CRK99432.1 EAA00982.5 KK107566 QOIP01000006 EZA48904.1 RLU21917.1 CH478006 EAT34447.1 JXUM01038855 JXUM01038856 KQ561182 KXJ79453.1 GL439972 EFN66448.1 GDHF01026788 JAI25526.1 GFDL01015416 JAV19629.1 DS233057 EDS27783.1 GBYB01000983 GBYB01000984 GBYB01000986 JAG70750.1 JAG70751.1 JAG70753.1 JRES01000586 KNC30034.1 LNIX01000001 OXA62139.1 OUUW01000035 SPP89828.1
NWSH01002297 PCG68611.1 ODYU01009703 SOQ54318.1 AGBW02010064 OWR49276.1 AGBW02010203 OWR48940.1 GDHF01001663 JAI50651.1 GAKP01009122 JAC49830.1 RSAL01000083 RVE48458.1 CH964239 EDW82304.1 GBXI01012330 GBXI01000453 JAD01962.1 JAD13839.1 JRES01001264 KNC24101.1 CCAG010000297 JXJN01012511 CH902621 EDV44538.1 CP012528 ALC49024.1 GL764255 EFZ18155.1 AE014298 BT031134 AY059453 AAL13359.1 ABX00756.1 GAMC01007563 GAMC01007562 JAB98993.1 CH480823 EDW56183.1 CH379063 EAL32554.2 CM000162 EDX02109.1 CH954180 EDV46725.1 KQ761783 OAD56995.1 ADMH02002023 ETN59944.1 GDAI01001857 JAI15746.1 GL888327 EGI62761.1 KZ288256 PBC30542.1 KQ980447 KYN16030.1 KQ976698 KYM77441.1 KQ976973 KYN06678.1 ADTU01015494 AAAB01008987 EGK97380.1 AXCN02000278 CH916371 EDV91774.1 APCN01002014 AXCM01000395 KQ981768 KYN35880.1 CH940655 EDW66248.1 GGFJ01005900 MBW55041.1 KQ982294 KYQ58359.1 GL445130 EFN90147.1 CH933812 EDW05872.1 ATLV01021913 KE525325 KFB46988.1 CVRI01000050 CRK99432.1 EAA00982.5 KK107566 QOIP01000006 EZA48904.1 RLU21917.1 CH478006 EAT34447.1 JXUM01038855 JXUM01038856 KQ561182 KXJ79453.1 GL439972 EFN66448.1 GDHF01026788 JAI25526.1 GFDL01015416 JAV19629.1 DS233057 EDS27783.1 GBYB01000983 GBYB01000984 GBYB01000986 JAG70750.1 JAG70751.1 JAG70753.1 JRES01000586 KNC30034.1 LNIX01000001 OXA62139.1 OUUW01000035 SPP89828.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000283053
+ More
UP000007798 UP000095300 UP000192221 UP000095301 UP000037069 UP000092445 UP000092444 UP000092460 UP000092443 UP000007801 UP000092553 UP000000803 UP000001292 UP000001819 UP000002282 UP000008711 UP000000673 UP000007755 UP000242457 UP000078492 UP000078540 UP000078542 UP000005203 UP000005205 UP000075885 UP000076407 UP000007062 UP000075886 UP000001070 UP000075840 UP000075883 UP000078541 UP000008792 UP000091820 UP000075881 UP000075880 UP000069272 UP000075882 UP000075809 UP000008237 UP000075884 UP000009192 UP000030765 UP000075903 UP000075900 UP000183832 UP000053097 UP000279307 UP000075901 UP000078200 UP000008820 UP000076408 UP000069940 UP000249989 UP000000311 UP000075920 UP000002320 UP000198287 UP000268350
UP000007798 UP000095300 UP000192221 UP000095301 UP000037069 UP000092445 UP000092444 UP000092460 UP000092443 UP000007801 UP000092553 UP000000803 UP000001292 UP000001819 UP000002282 UP000008711 UP000000673 UP000007755 UP000242457 UP000078492 UP000078540 UP000078542 UP000005203 UP000005205 UP000075885 UP000076407 UP000007062 UP000075886 UP000001070 UP000075840 UP000075883 UP000078541 UP000008792 UP000091820 UP000075881 UP000075880 UP000069272 UP000075882 UP000075809 UP000008237 UP000075884 UP000009192 UP000030765 UP000075903 UP000075900 UP000183832 UP000053097 UP000279307 UP000075901 UP000078200 UP000008820 UP000076408 UP000069940 UP000249989 UP000000311 UP000075920 UP000002320 UP000198287 UP000268350
Pfam
PF00782 DSPc
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
CDD
ProteinModelPortal
H9JEM0
A0A0N1IIT2
A0A194PW30
A0A2A4J9H9
A0A2H1WMR1
A0A212F6B2
+ More
A0A212F5D4 A0A0K8WHM0 A0A034W6R8 A0A437BDN4 B4NBU5 A0A0A1XRF2 A0A1I8Q332 A0A1W4VTG2 A0A1I8MD03 A0A0L0BVQ5 A0A1A9ZKU1 A0A1B0G8N9 A0A1B0BDL7 A0A1A9YMH8 B3MQB2 A0A0M5J635 E9IMS5 Q9VWF4 W8BQB5 B4I785 Q29IU9 B4PZF3 B3NVR0 A0A310SQ12 W5JB05 A0A0K8TNP7 F4WSV4 A0A2A3EGY1 A0A151J252 A0A195AZ10 A0A195D197 A0A088AW72 A0A158NH25 A0A182PNB8 A0A182XGH0 A0A1W5CAG9 A0A182QTS2 B4JM13 A0A182HZV6 A0A182M4P1 A0A195F5L2 B4MAI9 A0A1A9WVL4 A0A2M4BQ41 A0A182K5T2 A0A182JAV3 A0A182F7Z0 A0A182LQY6 A0A1S4GEJ6 A0A151XDD5 E2B2B0 A0A182N6A3 B4L676 A0A084W9U4 A0A182USS8 A0A182RT75 A0A1J1IGV1 Q7PYU3 A0A026W1G7 A0A182SKL9 A0A1A9UW41 Q16JJ4 A0A182XW25 A0A182G1Q7 E2AJC7 A0A182VRE3 A0A0K8UFZ6 A0A1Q3EWD7 B0XGR0 A0A0C9PJ41 A0A0L0CEX1 A0A226EX64 A0A3B0L0B5
A0A212F5D4 A0A0K8WHM0 A0A034W6R8 A0A437BDN4 B4NBU5 A0A0A1XRF2 A0A1I8Q332 A0A1W4VTG2 A0A1I8MD03 A0A0L0BVQ5 A0A1A9ZKU1 A0A1B0G8N9 A0A1B0BDL7 A0A1A9YMH8 B3MQB2 A0A0M5J635 E9IMS5 Q9VWF4 W8BQB5 B4I785 Q29IU9 B4PZF3 B3NVR0 A0A310SQ12 W5JB05 A0A0K8TNP7 F4WSV4 A0A2A3EGY1 A0A151J252 A0A195AZ10 A0A195D197 A0A088AW72 A0A158NH25 A0A182PNB8 A0A182XGH0 A0A1W5CAG9 A0A182QTS2 B4JM13 A0A182HZV6 A0A182M4P1 A0A195F5L2 B4MAI9 A0A1A9WVL4 A0A2M4BQ41 A0A182K5T2 A0A182JAV3 A0A182F7Z0 A0A182LQY6 A0A1S4GEJ6 A0A151XDD5 E2B2B0 A0A182N6A3 B4L676 A0A084W9U4 A0A182USS8 A0A182RT75 A0A1J1IGV1 Q7PYU3 A0A026W1G7 A0A182SKL9 A0A1A9UW41 Q16JJ4 A0A182XW25 A0A182G1Q7 E2AJC7 A0A182VRE3 A0A0K8UFZ6 A0A1Q3EWD7 B0XGR0 A0A0C9PJ41 A0A0L0CEX1 A0A226EX64 A0A3B0L0B5
PDB
6N8O
E-value=6.64941e-23,
Score=265
Ontologies
GO
Topology
Subcellular location
Nucleus
Mainly found in the nucleus. With evidence from 2 publications.
Cytoplasm Mainly found in the nucleus. With evidence from 2 publications.
Cytoplasm Mainly found in the nucleus. With evidence from 2 publications.
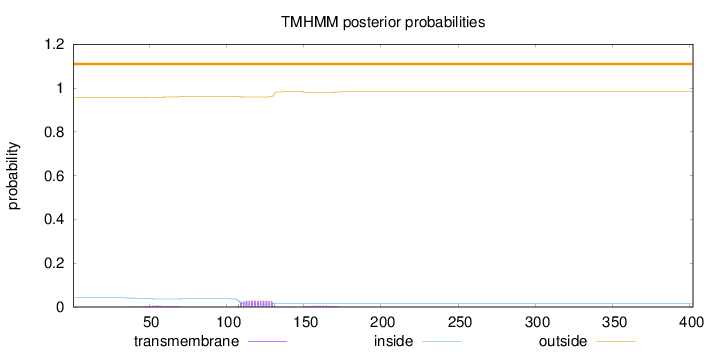
Length:
402
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.861129999999999
Exp number, first 60 AAs:
0.08914
Total prob of N-in:
0.04225
outside
1 - 402
Population Genetic Test Statistics
Pi
268.00354
Theta
182.586089
Tajima's D
1.561948
CLR
0.451874
CSRT
0.802109894505275
Interpretation
Uncertain
