Gene
KWMTBOMO08593
Pre Gene Modal
BGIBMGA007613
Annotation
Box_C/D_snoRNA_protein_1_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 3.816
Sequence
CDS
ATGTCTGCAGACTCAGACTGTGATGAACTGAGCCAAGATGGGCTTTCAAGACTTGGTGACTGCGAAGTTTGTGCGGCTAATAAAGCACTATACACATGTCCTAAATGCGAAGTTAAGACATGTTGCTTAAACTGCGTGAGAATCCACAAAAAAGAGTTGGAATGTGATGGTATAAGAGATCGGACAAAGTTTATAAGAATGAAAGATTTTACCGATACTGATCTCCTGAGTGACTACCGGCTGTTAGAAGAATGCGCGAGGTTTGTTTGGGGAGTCAAGAGAGACGATAAGAAGAGGTTCACAAGAATCGATAAGGATTTACCAATTCACCTATTCAAATTAAAAATGGCTGCACGCCAACGGGGTATTGTGCTTCAGTTTTTAGCACAAAATTTTACAAGACATAAAAATAACACTACAAGATACAATTATAAAAGTAACCTAATAAATTGGAGAGTCGAATGGATATTTCCTAATGTCGACAAAGAACCTATAAAATATGTTGATGAAAAGTGCCCTGAACAGAAAAAGCTATCTCATCTGTTGGATAAATATATTAACTGTGATGTGCCTTTTGAAGGATCAAAATCACTATCATTTTACAAGGCAGCTGGAATTAGTGGTCTTAAAATTTTACTTAAAGCCGAAAAAGTTAAAGGATCAGCCAAGAAATTCTTTGAACTAGATCCAAGTGAATCTTTAGCTGAGAATCTGTCTGGGAAATGTATTGTTGAATTTCCCATAATTTTCATTGTTTTAAAAGATCATGCATATAATTTTGAGATCATTACACCAGAAGACGAATTTGAAGAGATAGAAGCTAATAAACTTGTTAAAGAAATAAAACAAGATAATCATTCAGTTAATGGAATTACTAATGAAGAAAAATATATCAAAGAAAATCACAGAAAAAGACCAAGAGAATTATTAACAGAGAAAATTGCAAAAGAAAAAAGGATGGCATTAGAGAAGGAGATAAAAGAAGAACGAAAGAAGAAACCAAAAAATCTCTTGTTTACTACTGGATACTCATCTGAAGAAAGTCTTGAGGATAGTGATAATGAAAGTGAAACAGAAAAAAGTTGA
Protein
MSADSDCDELSQDGLSRLGDCEVCAANKALYTCPKCEVKTCCLNCVRIHKKELECDGIRDRTKFIRMKDFTDTDLLSDYRLLEECARFVWGVKRDDKKRFTRIDKDLPIHLFKLKMAARQRGIVLQFLAQNFTRHKNNTTRYNYKSNLINWRVEWIFPNVDKEPIKYVDEKCPEQKKLSHLLDKYINCDVPFEGSKSLSFYKAAGISGLKILLKAEKVKGSAKKFFELDPSESLAENLSGKCIVEFPIIFIVLKDHAYNFEIITPEDEFEEIEANKLVKEIKQDNHSVNGITNEEKYIKENHRKRPRELLTEKIAKEKRMALEKEIKEERKKKPKNLLFTTGYSSEESLEDSDNESETEKS
Summary
Uniprot
H9JDL6
A0A0L7LRD9
A0A194PQT5
A0A2A4JV77
A0A2H1V140
A0A437BDE7
+ More
A0A212F5E4 S4P096 A0A0N1IIT5 A0A2A3EFK7 A0A336MM05 Q16KT6 W8BR75 A0A1I8N4X2 A0A1I8NUS8 A0A1Q3G046 A0A0L0CC68 K7IRC2 A0A232ELU2 A0A088AK32 A0A034WGC9 A0A0A1X8K5 A0A084W182 E2BTD0 A0A0J7KRD4 A0A0K8W056 A0A1Y1M1X0 A0A151J148 A0A151WFU9 A0A182QUG7 A0A1A9XT04 A0A1A9ULF1 E2AEM7 A0A1B6C7I4 A0A195BSA9 A0A026X0S2 A0A158NV87 A0A151I8W1 A0A1B0D0J0 D6X4S3 E0W1V8 A0A182V2W1 A0A1B0CTM3 A0A182WAY6 A0A182ICU8 A0A182LLZ4 Q7Q8P2 A0A1B6JC86 A0A182FLP5 A0A067R4B5 A0A1B6FTZ1 A0A2J7QFT7 T1DSJ0 B4M879 A0A182TGI7 N6U5P4 B4JNJ1 J3JZB4 B4NDP5 A0A0P5FIF4 A0A164QEU8 A0A0P6GEL3 E9HV91 Q29J68 A0A0V0GCD7 A0A1J1HJ79 A0A146MHR1 A0A0A9Y997 A0A0P4X273 A0A443SPH0
A0A212F5E4 S4P096 A0A0N1IIT5 A0A2A3EFK7 A0A336MM05 Q16KT6 W8BR75 A0A1I8N4X2 A0A1I8NUS8 A0A1Q3G046 A0A0L0CC68 K7IRC2 A0A232ELU2 A0A088AK32 A0A034WGC9 A0A0A1X8K5 A0A084W182 E2BTD0 A0A0J7KRD4 A0A0K8W056 A0A1Y1M1X0 A0A151J148 A0A151WFU9 A0A182QUG7 A0A1A9XT04 A0A1A9ULF1 E2AEM7 A0A1B6C7I4 A0A195BSA9 A0A026X0S2 A0A158NV87 A0A151I8W1 A0A1B0D0J0 D6X4S3 E0W1V8 A0A182V2W1 A0A1B0CTM3 A0A182WAY6 A0A182ICU8 A0A182LLZ4 Q7Q8P2 A0A1B6JC86 A0A182FLP5 A0A067R4B5 A0A1B6FTZ1 A0A2J7QFT7 T1DSJ0 B4M879 A0A182TGI7 N6U5P4 B4JNJ1 J3JZB4 B4NDP5 A0A0P5FIF4 A0A164QEU8 A0A0P6GEL3 E9HV91 Q29J68 A0A0V0GCD7 A0A1J1HJ79 A0A146MHR1 A0A0A9Y997 A0A0P4X273 A0A443SPH0
Pubmed
19121390
26227816
26354079
22118469
23622113
17510324
+ More
24495485 25315136 26108605 20075255 28648823 25348373 25830018 24438588 20798317 28004739 24508170 30249741 21347285 18362917 19820115 20566863 20966253 12364791 14747013 17210077 24845553 17994087 23537049 22516182 21292972 15632085 26823975 25401762
24495485 25315136 26108605 20075255 28648823 25348373 25830018 24438588 20798317 28004739 24508170 30249741 21347285 18362917 19820115 20566863 20966253 12364791 14747013 17210077 24845553 17994087 23537049 22516182 21292972 15632085 26823975 25401762
EMBL
BABH01003324
JTDY01000260
KOB78027.1
KQ459596
KPI95323.1
NWSH01000517
+ More
PCG75911.1 ODYU01000192 SOQ34565.1 RSAL01000083 RVE48449.1 AGBW02010203 OWR48955.1 GAIX01011007 JAA81553.1 KQ459717 KPJ20304.1 KZ288256 PBC30573.1 UFQT01001695 SSX31454.1 CH477938 EAT34923.1 EJY58005.1 GAMC01007222 GAMC01007220 JAB99333.1 GFDL01001854 JAV33191.1 JRES01000619 KNC29841.1 NNAY01003494 OXU19333.1 GAKP01004316 JAC54636.1 GBXI01007289 JAD07003.1 ATLV01019244 KE525266 KFB43976.1 GL450389 EFN81051.1 LBMM01003973 KMQ92937.1 GDHF01007771 JAI44543.1 GEZM01044290 JAV78325.1 KQ980575 KYN15497.1 KQ983203 KYQ46717.1 AXCN02001891 GL438870 EFN68102.1 GEDC01027845 JAS09453.1 KQ976417 KYM89905.1 KK107039 QOIP01000006 EZA61905.1 RLU21250.1 ADTU01027002 KQ978330 KYM95085.1 AJVK01009949 KQ971380 EEZ97683.1 DS235873 EEB19552.1 AJWK01027780 APCN01008106 AAAB01008942 EAA10029.2 GECU01010978 JAS96728.1 KK852705 KDR18016.1 GECZ01016316 JAS53453.1 NEVH01014842 PNF27456.1 GAMD01000170 JAB01421.1 CH940653 EDW62355.2 APGK01047873 KB741092 KB632050 ENN73907.1 ERL88299.1 CH916371 EDV92284.1 BT128595 AEE63552.1 CH964239 EDW81867.2 GDIQ01256721 JAJ95003.1 LRGB01002443 KZS07688.1 GDIQ01034231 JAN60506.1 GL732844 EFX64345.1 CH379063 EAL32434.2 GECL01001069 JAP05055.1 CVRI01000001 CRK86526.1 GDHC01000067 JAQ18562.1 GBHO01042866 GBHO01042864 GBHO01032340 GBHO01014829 GBHO01014828 GBHO01014827 GBHO01014826 GBHO01014825 GBHO01014824 GBHO01003726 GBRD01006548 GBRD01001476 JAG00738.1 JAG00740.1 JAG11264.1 JAG28775.1 JAG28776.1 JAG28777.1 JAG28778.1 JAG28779.1 JAG28780.1 JAG39878.1 JAG59273.1 GDRN01010858 JAI67908.1 NCKV01000921 RWS29428.1
PCG75911.1 ODYU01000192 SOQ34565.1 RSAL01000083 RVE48449.1 AGBW02010203 OWR48955.1 GAIX01011007 JAA81553.1 KQ459717 KPJ20304.1 KZ288256 PBC30573.1 UFQT01001695 SSX31454.1 CH477938 EAT34923.1 EJY58005.1 GAMC01007222 GAMC01007220 JAB99333.1 GFDL01001854 JAV33191.1 JRES01000619 KNC29841.1 NNAY01003494 OXU19333.1 GAKP01004316 JAC54636.1 GBXI01007289 JAD07003.1 ATLV01019244 KE525266 KFB43976.1 GL450389 EFN81051.1 LBMM01003973 KMQ92937.1 GDHF01007771 JAI44543.1 GEZM01044290 JAV78325.1 KQ980575 KYN15497.1 KQ983203 KYQ46717.1 AXCN02001891 GL438870 EFN68102.1 GEDC01027845 JAS09453.1 KQ976417 KYM89905.1 KK107039 QOIP01000006 EZA61905.1 RLU21250.1 ADTU01027002 KQ978330 KYM95085.1 AJVK01009949 KQ971380 EEZ97683.1 DS235873 EEB19552.1 AJWK01027780 APCN01008106 AAAB01008942 EAA10029.2 GECU01010978 JAS96728.1 KK852705 KDR18016.1 GECZ01016316 JAS53453.1 NEVH01014842 PNF27456.1 GAMD01000170 JAB01421.1 CH940653 EDW62355.2 APGK01047873 KB741092 KB632050 ENN73907.1 ERL88299.1 CH916371 EDV92284.1 BT128595 AEE63552.1 CH964239 EDW81867.2 GDIQ01256721 JAJ95003.1 LRGB01002443 KZS07688.1 GDIQ01034231 JAN60506.1 GL732844 EFX64345.1 CH379063 EAL32434.2 GECL01001069 JAP05055.1 CVRI01000001 CRK86526.1 GDHC01000067 JAQ18562.1 GBHO01042866 GBHO01042864 GBHO01032340 GBHO01014829 GBHO01014828 GBHO01014827 GBHO01014826 GBHO01014825 GBHO01014824 GBHO01003726 GBRD01006548 GBRD01001476 JAG00738.1 JAG00740.1 JAG11264.1 JAG28775.1 JAG28776.1 JAG28777.1 JAG28778.1 JAG28779.1 JAG28780.1 JAG39878.1 JAG59273.1 GDRN01010858 JAI67908.1 NCKV01000921 RWS29428.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000218220
UP000283053
UP000007151
+ More
UP000053240 UP000242457 UP000008820 UP000095301 UP000095300 UP000037069 UP000002358 UP000215335 UP000005203 UP000030765 UP000008237 UP000036403 UP000078492 UP000075809 UP000075886 UP000092443 UP000078200 UP000000311 UP000078540 UP000053097 UP000279307 UP000005205 UP000078542 UP000092462 UP000007266 UP000009046 UP000075903 UP000092461 UP000075920 UP000075840 UP000075882 UP000007062 UP000069272 UP000027135 UP000235965 UP000008792 UP000075902 UP000019118 UP000030742 UP000001070 UP000007798 UP000076858 UP000000305 UP000001819 UP000183832 UP000288716
UP000053240 UP000242457 UP000008820 UP000095301 UP000095300 UP000037069 UP000002358 UP000215335 UP000005203 UP000030765 UP000008237 UP000036403 UP000078492 UP000075809 UP000075886 UP000092443 UP000078200 UP000000311 UP000078540 UP000053097 UP000279307 UP000005205 UP000078542 UP000092462 UP000007266 UP000009046 UP000075903 UP000092461 UP000075920 UP000075840 UP000075882 UP000007062 UP000069272 UP000027135 UP000235965 UP000008792 UP000075902 UP000019118 UP000030742 UP000001070 UP000007798 UP000076858 UP000000305 UP000001819 UP000183832 UP000288716
PRIDE
Pfam
PF04438 zf-HIT
CDD
ProteinModelPortal
H9JDL6
A0A0L7LRD9
A0A194PQT5
A0A2A4JV77
A0A2H1V140
A0A437BDE7
+ More
A0A212F5E4 S4P096 A0A0N1IIT5 A0A2A3EFK7 A0A336MM05 Q16KT6 W8BR75 A0A1I8N4X2 A0A1I8NUS8 A0A1Q3G046 A0A0L0CC68 K7IRC2 A0A232ELU2 A0A088AK32 A0A034WGC9 A0A0A1X8K5 A0A084W182 E2BTD0 A0A0J7KRD4 A0A0K8W056 A0A1Y1M1X0 A0A151J148 A0A151WFU9 A0A182QUG7 A0A1A9XT04 A0A1A9ULF1 E2AEM7 A0A1B6C7I4 A0A195BSA9 A0A026X0S2 A0A158NV87 A0A151I8W1 A0A1B0D0J0 D6X4S3 E0W1V8 A0A182V2W1 A0A1B0CTM3 A0A182WAY6 A0A182ICU8 A0A182LLZ4 Q7Q8P2 A0A1B6JC86 A0A182FLP5 A0A067R4B5 A0A1B6FTZ1 A0A2J7QFT7 T1DSJ0 B4M879 A0A182TGI7 N6U5P4 B4JNJ1 J3JZB4 B4NDP5 A0A0P5FIF4 A0A164QEU8 A0A0P6GEL3 E9HV91 Q29J68 A0A0V0GCD7 A0A1J1HJ79 A0A146MHR1 A0A0A9Y997 A0A0P4X273 A0A443SPH0
A0A212F5E4 S4P096 A0A0N1IIT5 A0A2A3EFK7 A0A336MM05 Q16KT6 W8BR75 A0A1I8N4X2 A0A1I8NUS8 A0A1Q3G046 A0A0L0CC68 K7IRC2 A0A232ELU2 A0A088AK32 A0A034WGC9 A0A0A1X8K5 A0A084W182 E2BTD0 A0A0J7KRD4 A0A0K8W056 A0A1Y1M1X0 A0A151J148 A0A151WFU9 A0A182QUG7 A0A1A9XT04 A0A1A9ULF1 E2AEM7 A0A1B6C7I4 A0A195BSA9 A0A026X0S2 A0A158NV87 A0A151I8W1 A0A1B0D0J0 D6X4S3 E0W1V8 A0A182V2W1 A0A1B0CTM3 A0A182WAY6 A0A182ICU8 A0A182LLZ4 Q7Q8P2 A0A1B6JC86 A0A182FLP5 A0A067R4B5 A0A1B6FTZ1 A0A2J7QFT7 T1DSJ0 B4M879 A0A182TGI7 N6U5P4 B4JNJ1 J3JZB4 B4NDP5 A0A0P5FIF4 A0A164QEU8 A0A0P6GEL3 E9HV91 Q29J68 A0A0V0GCD7 A0A1J1HJ79 A0A146MHR1 A0A0A9Y997 A0A0P4X273 A0A443SPH0
PDB
2N94
E-value=0.00758797,
Score=92
Ontologies
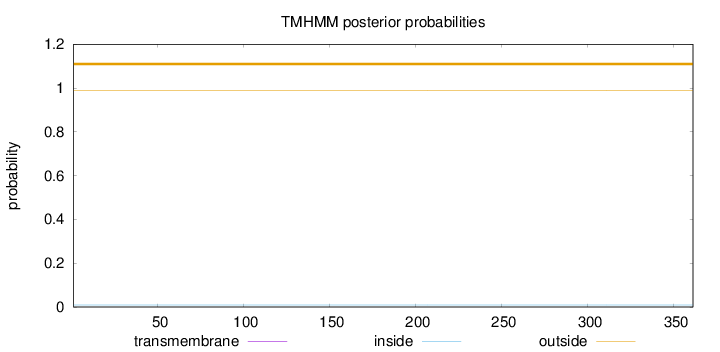
Topology
Length:
361
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00181
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00954
outside
1 - 361
Population Genetic Test Statistics
Pi
145.047625
Theta
133.311969
Tajima's D
0.148941
CLR
0.693226
CSRT
0.409929503524824
Interpretation
Uncertain
