Pre Gene Modal
BGIBMGA007973
Annotation
PREDICTED:_PAXIP1-associated_glutamate-rich_protein_1_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.586
Sequence
CDS
ATGGAAACCGAGATTCAAGGAGACGATTGGGATATCCCTTGTTCAGATGACGAACTGGCGAAAAACGGCGTTTTTATTGGAAAAAGCTGGATGCCGGAACCGGCCGAATTAGAGGAACTTTACAAAAACATCGACAAAAAGGGTATATTAAACCTAGAATGGAAATGTCCAGGACGCAGGCAACCATCTCCCGTGCCTGTCAGCAAGGAAAACCAGCCTGAGTTACCTCAATCGCCAATCCGGGAAGAAAAGTCAGACTTTGATTTCATGGATGAGGTTAGTTCATTGCGGCTGCGGGTAAGACGGGAAGGGGAAGACACGTTGAGAGGTTCTGCTAAGAAAAAGACAACTTCTTTTGATGGTATTCTTTCGAATATGATAAGGCATAAAAGAATAGAGCAGATGGAAAAACAAGCGAAAACATCACCCTCAACTACTTCAAGTAACTAA
Protein
METEIQGDDWDIPCSDDELAKNGVFIGKSWMPEPAELEELYKNIDKKGILNLEWKCPGRRQPSPVPVSKENQPELPQSPIREEKSDFDFMDEVSSLRLRVRREGEDTLRGSAKKKTTSFDGILSNMIRHKRIEQMEKQAKTSPSTTSSN
Summary
Uniprot
H9JEM6
A0A437BDM4
A0A0N1ID16
A0A0L7LRJ6
A0A2A4JWZ3
A0A194PPT1
+ More
A0A2H1V2R8 A0A212F5E3 S4PLW9 A0A2J7Q2P9 A0A347ZJ88 A0A2P8Z332 A0A0M9A1L9 E2A3W1 A0A0J7KI48 A0A310SM60 A0A2A3ENN0 A0A087ZP49 A0A067QS70 A0A026WU19 A0A0L7RAA3 A0A232EFV1 E2BYL1 A0A1W4XHX3 A0A154PTT5 A0A195EM04 E9IWW1 A0A195ETN8 A0A1Y1MJ53 A0A158NPP9 A0A151X8N8 A0A195ATQ2 A0A1B6KC38 U4UJR3 A0A1B6CU17 A0A0T6B2A4 N6TVU7 A0A0C9R6G3 A0A2R7WC67 A0A1B6JE64 J9JKX9 A0A023F0Z4 A0A1B6F256 T1HZA4 A0A0V0GES7 A0A069DVX6 A0A453YJV2 A0A182UXS1 A0A182UB88 A0A182KWR7 A0A182XKC8 Q7Q9A7 A0A182XWM4 A0A182KGS9 A0A182Q6R5 A0A182MNT5 A0A182RDA1 A0A146M054 A0A0A9W8J3 A0A182VY93 A0A084VUZ0 A0A182JBX9 T1INR3 A0A182NSS0 E9HWW2 A0A0P6AB26 A0A0P5ZJF4 W5JDJ4 A0A0P5FXA7 A0A182F7W0 A0A2M3ZKA5 A0A146L2X0 A0A0P4VQN8 A0A224XP48 A0A087USF6 A0A1Q3FBS0 A0A2L2YF13 A0A0P4W9H3 A0A0P6IUJ7 Q16P10 A0A1L8DAG7 A0A0P5CHT3 A0A170VV25 A0A182PX16 T1E7R8 B0X6A1 A0A1W7R524 A0A1L8DB72 A0A1L8DB16 A0A1B0DGQ5 V5HF87 B7P813 U5EN78
A0A2H1V2R8 A0A212F5E3 S4PLW9 A0A2J7Q2P9 A0A347ZJ88 A0A2P8Z332 A0A0M9A1L9 E2A3W1 A0A0J7KI48 A0A310SM60 A0A2A3ENN0 A0A087ZP49 A0A067QS70 A0A026WU19 A0A0L7RAA3 A0A232EFV1 E2BYL1 A0A1W4XHX3 A0A154PTT5 A0A195EM04 E9IWW1 A0A195ETN8 A0A1Y1MJ53 A0A158NPP9 A0A151X8N8 A0A195ATQ2 A0A1B6KC38 U4UJR3 A0A1B6CU17 A0A0T6B2A4 N6TVU7 A0A0C9R6G3 A0A2R7WC67 A0A1B6JE64 J9JKX9 A0A023F0Z4 A0A1B6F256 T1HZA4 A0A0V0GES7 A0A069DVX6 A0A453YJV2 A0A182UXS1 A0A182UB88 A0A182KWR7 A0A182XKC8 Q7Q9A7 A0A182XWM4 A0A182KGS9 A0A182Q6R5 A0A182MNT5 A0A182RDA1 A0A146M054 A0A0A9W8J3 A0A182VY93 A0A084VUZ0 A0A182JBX9 T1INR3 A0A182NSS0 E9HWW2 A0A0P6AB26 A0A0P5ZJF4 W5JDJ4 A0A0P5FXA7 A0A182F7W0 A0A2M3ZKA5 A0A146L2X0 A0A0P4VQN8 A0A224XP48 A0A087USF6 A0A1Q3FBS0 A0A2L2YF13 A0A0P4W9H3 A0A0P6IUJ7 Q16P10 A0A1L8DAG7 A0A0P5CHT3 A0A170VV25 A0A182PX16 T1E7R8 B0X6A1 A0A1W7R524 A0A1L8DB72 A0A1L8DB16 A0A1B0DGQ5 V5HF87 B7P813 U5EN78
Pubmed
19121390
26354079
26227816
22118469
23622113
29403074
+ More
20798317 24845553 24508170 30249741 28648823 21282665 28004739 21347285 23537049 25474469 26334808 20966253 12364791 14747013 17210077 25244985 26823975 25401762 24438588 21292972 20920257 23761445 27129103 26561354 26999592 17510324 25765539
20798317 24845553 24508170 30249741 28648823 21282665 28004739 21347285 23537049 25474469 26334808 20966253 12364791 14747013 17210077 25244985 26823975 25401762 24438588 21292972 20920257 23761445 27129103 26561354 26999592 17510324 25765539
EMBL
BABH01003324
RSAL01000083
RVE48448.1
KQ459717
KPJ20305.1
JTDY01000260
+ More
KOB78024.1 NWSH01000517 PCG75912.1 KQ459596 KPI95322.1 ODYU01000192 SOQ34564.1 AGBW02010203 OWR48956.1 GAIX01003820 JAA88740.1 NEVH01019092 PNF22854.1 FX985656 BBA84394.1 PYGN01000219 PSN50912.1 KQ435784 KOX74546.1 GL436510 EFN71885.1 LBMM01007243 KMQ89914.1 KQ761343 OAD57865.1 KZ288204 PBC33327.1 KK853090 KDR11568.1 KK107105 QOIP01000011 EZA59522.1 RLU16636.1 KQ414619 KOC67774.1 NNAY01004934 OXU17214.1 GL451493 EFN79216.1 KQ435090 KZC14610.1 KQ978730 KYN28952.1 GL766616 EFZ14895.1 KQ981986 KYN31244.1 GEZM01032339 JAV84660.1 ADTU01022705 KQ982409 KYQ56680.1 KQ976745 KYM75432.1 GEBQ01030960 JAT09017.1 KB632353 ERL93327.1 GEDC01026558 GEDC01023744 GEDC01023477 GEDC01020575 GEDC01016525 GEDC01015083 GEDC01014917 JAS10740.1 JAS13554.1 JAS13821.1 JAS16723.1 JAS20773.1 JAS22215.1 JAS22381.1 LJIG01016150 KRT81465.1 APGK01050089 KB741165 ENN73420.1 GBYB01002351 GBYB01002352 JAG72118.1 JAG72119.1 KK854557 PTY16831.1 GECU01010287 JAS97419.1 ABLF02019206 GBBI01003769 JAC14943.1 GECZ01025484 JAS44285.1 ACPB03014184 GECL01000317 JAP05807.1 GBGD01003435 JAC85454.1 APCN01002180 AAAB01008904 EAA09607.1 AXCN02000227 AXCM01018426 GDHC01006639 JAQ11990.1 GBHO01042460 GBHO01042459 GBHO01042458 GBHO01042456 GBHO01042455 GBRD01007660 GDHC01001307 JAG01144.1 JAG01145.1 JAG01146.1 JAG01148.1 JAG01149.1 JAG58161.1 JAQ17322.1 ATLV01017129 KE525156 KFB41784.1 JH431198 GL732966 EFX63770.1 GDIP01032148 JAM71567.1 GDIP01043105 JAM60610.1 ADMH02001528 ETN62171.1 GDIQ01256194 JAJ95530.1 GGFM01008171 MBW28922.1 GDHC01016973 JAQ01656.1 GDKW01003122 JAI53473.1 GFTR01002170 JAW14256.1 KK121351 KFM80295.1 GFDL01010086 JAV24959.1 IAAA01026187 IAAA01026188 LAA06709.1 GDRN01062374 JAI65081.1 GDUN01000485 JAN95434.1 CH477800 EAT36091.1 GFDF01010662 JAV03422.1 GDIP01171206 JAJ52196.1 GEMB01006372 JAR96966.1 GAMD01002817 JAA98773.1 DS232407 EDS41251.1 GEHC01001386 JAV46259.1 GFDF01010376 JAV03708.1 GFDF01010421 JAV03663.1 AJVK01033811 GANP01002173 JAB82295.1 ABJB010133960 DS654682 EEC02735.1 GANO01004227 JAB55644.1
KOB78024.1 NWSH01000517 PCG75912.1 KQ459596 KPI95322.1 ODYU01000192 SOQ34564.1 AGBW02010203 OWR48956.1 GAIX01003820 JAA88740.1 NEVH01019092 PNF22854.1 FX985656 BBA84394.1 PYGN01000219 PSN50912.1 KQ435784 KOX74546.1 GL436510 EFN71885.1 LBMM01007243 KMQ89914.1 KQ761343 OAD57865.1 KZ288204 PBC33327.1 KK853090 KDR11568.1 KK107105 QOIP01000011 EZA59522.1 RLU16636.1 KQ414619 KOC67774.1 NNAY01004934 OXU17214.1 GL451493 EFN79216.1 KQ435090 KZC14610.1 KQ978730 KYN28952.1 GL766616 EFZ14895.1 KQ981986 KYN31244.1 GEZM01032339 JAV84660.1 ADTU01022705 KQ982409 KYQ56680.1 KQ976745 KYM75432.1 GEBQ01030960 JAT09017.1 KB632353 ERL93327.1 GEDC01026558 GEDC01023744 GEDC01023477 GEDC01020575 GEDC01016525 GEDC01015083 GEDC01014917 JAS10740.1 JAS13554.1 JAS13821.1 JAS16723.1 JAS20773.1 JAS22215.1 JAS22381.1 LJIG01016150 KRT81465.1 APGK01050089 KB741165 ENN73420.1 GBYB01002351 GBYB01002352 JAG72118.1 JAG72119.1 KK854557 PTY16831.1 GECU01010287 JAS97419.1 ABLF02019206 GBBI01003769 JAC14943.1 GECZ01025484 JAS44285.1 ACPB03014184 GECL01000317 JAP05807.1 GBGD01003435 JAC85454.1 APCN01002180 AAAB01008904 EAA09607.1 AXCN02000227 AXCM01018426 GDHC01006639 JAQ11990.1 GBHO01042460 GBHO01042459 GBHO01042458 GBHO01042456 GBHO01042455 GBRD01007660 GDHC01001307 JAG01144.1 JAG01145.1 JAG01146.1 JAG01148.1 JAG01149.1 JAG58161.1 JAQ17322.1 ATLV01017129 KE525156 KFB41784.1 JH431198 GL732966 EFX63770.1 GDIP01032148 JAM71567.1 GDIP01043105 JAM60610.1 ADMH02001528 ETN62171.1 GDIQ01256194 JAJ95530.1 GGFM01008171 MBW28922.1 GDHC01016973 JAQ01656.1 GDKW01003122 JAI53473.1 GFTR01002170 JAW14256.1 KK121351 KFM80295.1 GFDL01010086 JAV24959.1 IAAA01026187 IAAA01026188 LAA06709.1 GDRN01062374 JAI65081.1 GDUN01000485 JAN95434.1 CH477800 EAT36091.1 GFDF01010662 JAV03422.1 GDIP01171206 JAJ52196.1 GEMB01006372 JAR96966.1 GAMD01002817 JAA98773.1 DS232407 EDS41251.1 GEHC01001386 JAV46259.1 GFDF01010376 JAV03708.1 GFDF01010421 JAV03663.1 AJVK01033811 GANP01002173 JAB82295.1 ABJB010133960 DS654682 EEC02735.1 GANO01004227 JAB55644.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000037510
UP000218220
UP000053268
+ More
UP000007151 UP000235965 UP000245037 UP000053105 UP000000311 UP000036403 UP000242457 UP000005203 UP000027135 UP000053097 UP000279307 UP000053825 UP000215335 UP000008237 UP000192223 UP000076502 UP000078492 UP000078541 UP000005205 UP000075809 UP000078540 UP000030742 UP000019118 UP000007819 UP000015103 UP000075840 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000076408 UP000075881 UP000075886 UP000075883 UP000075900 UP000075920 UP000030765 UP000075880 UP000075884 UP000000305 UP000000673 UP000069272 UP000054359 UP000008820 UP000075885 UP000002320 UP000092462 UP000001555
UP000007151 UP000235965 UP000245037 UP000053105 UP000000311 UP000036403 UP000242457 UP000005203 UP000027135 UP000053097 UP000279307 UP000053825 UP000215335 UP000008237 UP000192223 UP000076502 UP000078492 UP000078541 UP000005205 UP000075809 UP000078540 UP000030742 UP000019118 UP000007819 UP000015103 UP000075840 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000076408 UP000075881 UP000075886 UP000075883 UP000075900 UP000075920 UP000030765 UP000075880 UP000075884 UP000000305 UP000000673 UP000069272 UP000054359 UP000008820 UP000075885 UP000002320 UP000092462 UP000001555
PRIDE
Pfam
PF15364 PAXIP1_C
Interpro
IPR028213
PA1
ProteinModelPortal
H9JEM6
A0A437BDM4
A0A0N1ID16
A0A0L7LRJ6
A0A2A4JWZ3
A0A194PPT1
+ More
A0A2H1V2R8 A0A212F5E3 S4PLW9 A0A2J7Q2P9 A0A347ZJ88 A0A2P8Z332 A0A0M9A1L9 E2A3W1 A0A0J7KI48 A0A310SM60 A0A2A3ENN0 A0A087ZP49 A0A067QS70 A0A026WU19 A0A0L7RAA3 A0A232EFV1 E2BYL1 A0A1W4XHX3 A0A154PTT5 A0A195EM04 E9IWW1 A0A195ETN8 A0A1Y1MJ53 A0A158NPP9 A0A151X8N8 A0A195ATQ2 A0A1B6KC38 U4UJR3 A0A1B6CU17 A0A0T6B2A4 N6TVU7 A0A0C9R6G3 A0A2R7WC67 A0A1B6JE64 J9JKX9 A0A023F0Z4 A0A1B6F256 T1HZA4 A0A0V0GES7 A0A069DVX6 A0A453YJV2 A0A182UXS1 A0A182UB88 A0A182KWR7 A0A182XKC8 Q7Q9A7 A0A182XWM4 A0A182KGS9 A0A182Q6R5 A0A182MNT5 A0A182RDA1 A0A146M054 A0A0A9W8J3 A0A182VY93 A0A084VUZ0 A0A182JBX9 T1INR3 A0A182NSS0 E9HWW2 A0A0P6AB26 A0A0P5ZJF4 W5JDJ4 A0A0P5FXA7 A0A182F7W0 A0A2M3ZKA5 A0A146L2X0 A0A0P4VQN8 A0A224XP48 A0A087USF6 A0A1Q3FBS0 A0A2L2YF13 A0A0P4W9H3 A0A0P6IUJ7 Q16P10 A0A1L8DAG7 A0A0P5CHT3 A0A170VV25 A0A182PX16 T1E7R8 B0X6A1 A0A1W7R524 A0A1L8DB72 A0A1L8DB16 A0A1B0DGQ5 V5HF87 B7P813 U5EN78
A0A2H1V2R8 A0A212F5E3 S4PLW9 A0A2J7Q2P9 A0A347ZJ88 A0A2P8Z332 A0A0M9A1L9 E2A3W1 A0A0J7KI48 A0A310SM60 A0A2A3ENN0 A0A087ZP49 A0A067QS70 A0A026WU19 A0A0L7RAA3 A0A232EFV1 E2BYL1 A0A1W4XHX3 A0A154PTT5 A0A195EM04 E9IWW1 A0A195ETN8 A0A1Y1MJ53 A0A158NPP9 A0A151X8N8 A0A195ATQ2 A0A1B6KC38 U4UJR3 A0A1B6CU17 A0A0T6B2A4 N6TVU7 A0A0C9R6G3 A0A2R7WC67 A0A1B6JE64 J9JKX9 A0A023F0Z4 A0A1B6F256 T1HZA4 A0A0V0GES7 A0A069DVX6 A0A453YJV2 A0A182UXS1 A0A182UB88 A0A182KWR7 A0A182XKC8 Q7Q9A7 A0A182XWM4 A0A182KGS9 A0A182Q6R5 A0A182MNT5 A0A182RDA1 A0A146M054 A0A0A9W8J3 A0A182VY93 A0A084VUZ0 A0A182JBX9 T1INR3 A0A182NSS0 E9HWW2 A0A0P6AB26 A0A0P5ZJF4 W5JDJ4 A0A0P5FXA7 A0A182F7W0 A0A2M3ZKA5 A0A146L2X0 A0A0P4VQN8 A0A224XP48 A0A087USF6 A0A1Q3FBS0 A0A2L2YF13 A0A0P4W9H3 A0A0P6IUJ7 Q16P10 A0A1L8DAG7 A0A0P5CHT3 A0A170VV25 A0A182PX16 T1E7R8 B0X6A1 A0A1W7R524 A0A1L8DB72 A0A1L8DB16 A0A1B0DGQ5 V5HF87 B7P813 U5EN78
Ontologies
KEGG
PANTHER
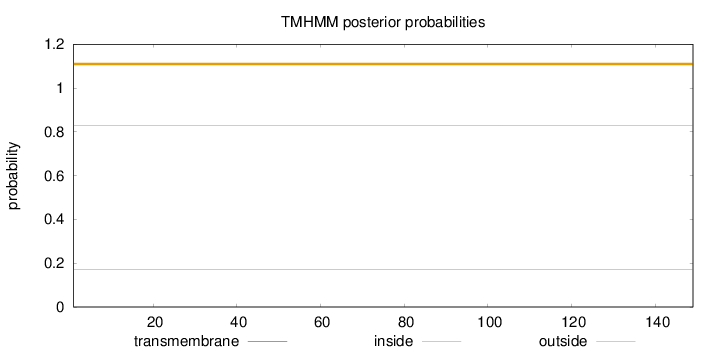
Topology
Length:
149
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.17199
outside
1 - 149
Population Genetic Test Statistics
Pi
149.739494
Theta
144.808268
Tajima's D
0.160911
CLR
0.01613
CSRT
0.415329233538323
Interpretation
Uncertain
