Gene
KWMTBOMO08589
Pre Gene Modal
BGIBMGA007609
Annotation
PREDICTED:_protein_crumbs_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.056
Sequence
CDS
ATGGCACACGTAAAATTGGTTTTCATGTTTTTGTTTGTCAAAGGTAGATATTGCGAGACAGACGTTGACGAATGCGCCATGAAGCCTAAGATATGCAACAACGGGGTTTGTGTGAACACACCAGGCTCCTACCAGTGTTATTGTAGGCCAGGATATACGGGCGATAGCTGCGAGCAAGACATCGACGAATGTCTCTCGTCGCCGTGCAAAAACAGCGGCACATGTCAGAACTTGGAGAATAATTACGAATGCGTTTGTATGGAGGGCTTTGAAGGTAAAGATTGTTCGGTCAACATCGACGAGTGCGAGGCGTCCCCGTGCGCGGCGGGCTCCACGTGCGTGGACGGCGTGGCCTCCTACTCCTGCCAGTGCCAGGAGGGGCTCACCGGCCCGCGCTGTGAGGTCGACATCGACGACTGCGAGTCGCAGCCGTGCCAGCACGGGGGCCGCTGCGTGGACGCGCTCAACGGCTACAGCTGCGAGTGCGGCGGCACCGGCTTCACCGGCGACGACTGCTCCGTCAACATCGACGAGTGCGCGCCCGCGCCCTGCCGCCACGGAGCCACCTGCGTGGACGAGCTCAACGACTACCGCTGCTCCTGCCACCCCGGCTTCACGGGCAAGAACTGCGAGGTGGACATCAACGAGTGCGAGGGCGAGCCCTGCAAGTTCGGCGGCGTGTGTCTGGAGCGCAGCAACGCCAGCCTGTACCTGGCGCCCGACGCGCCGCCCGTGCTGGTGGGCCCGCACGACGCCATGCTGCTGCCGCCCGCCTTCTACCAGCCCTTCGCCATCGACGACGCCAGCGGGTTCGAGTGCGTGTGCGTGCCGGGCACGGCGGGCGCGCGCTGCGAGCTCAACATCGACGAGTGCGCGTCGTCCCCGTGCGGACACGGCAAGTGCGTGGACGGCGTGGGCTCCTACTCCTGCGACTGCCTGCCGGGCTACGAGGGGGACCACTGCGAGATCGAAATCGACGAGTGCCAGAGGTACCAGCCGTGCGCGCACGGGCGCTGCGTGGACCAGCGCGCCACGTACCACTGCTCGTGCTCGGCGGGCTGGGGCGGCCGGAACTGCTCCGTGGTGCTGCAGGGCTGCGCCGCGCAACCCTGCCGCAACCAGGGCTCCTGCCACCCCTGGCTGCGCGACGAGACCGAGCACCGCTTCAACTGCTCCTGCGCCAACGGCTTCTACGGCACCACCTGCGAGAAGATCACCACCATGTCCCTGGAGAGGAGCAGCTACGCGGAGGTGAACACTTCGCGCGAGGAGGGCTACGACATCTCGTTCCGCTTCAAGACGACGCTGGGCGACGGGCTGCTGGCCATGGGCATCGGCCTCACCTACTTCTTCCTCGAGCTGTCCAACGGCCGCCTCAACCTGCACTCCAGCCTGCTCAACAAGTGGGAGGGCGTCTTCATCGGCTCCAACCTCAACGACAGCAACTGGCAGAAGGTGTTCGTGACGATAAACTCCTCGCACCTGGTGTTGGCCGCCAACGAAGAGCAGACCATCTACCCCATCAGCCAGATCGAAGCGTACAACTCGTCGGTGACGTCGTTCCCCACCACGCGGCTCGGGAGGGCCGGCTCCTCCTTCGCGACCCTCACCCACGGACCTGACTTCTTCGTCGGCTGCTTCCAAGATGTCGTCGTGAACGGACAGTGGGTGCTGCCGGAGAGCGGAGAGAGCGAGGGCGAGGGCCCGGTGTGGCGCCTGGCGGGCGTGACGGCGGCGTGTCCCCGCGTGCCGCAGTGCCAGCCCTCCCCCTGCCGCTCGGGGGGGCGCTGCCACGACCACTGGACCTCCTTCCTGTGCACCTGCCCGCGGCCGCACCTGGGAGACACCTGCCAGTACAACTACACGGCGGCGACGTTCGGGCACGAGGCGGCGCGGCGCGGCTCGGCGGTGCGCGTGCTGGTGGGGGAGGCGGCTCGGCGCGCCGTGCTGGCCGCGCTCGACATCTCCATGTTCATCCGGACCAGGAAACCCACCGGACAGATCTTTTACTTGGGCTCTCTGCCAAGGTACGGGCAGACCGAGGAGACGCAGGTGGGCGCCTCGCTGTCCAACGGCGAGCTGCTGGTGCACCTGCGCTTCAACCAGACCCCGGAGAACTACACCGTGGGCGGCACCCGCCTGGACAACGGACACCTGCACCTCATCGAGGTGGTCCGCAACTCGACGTTAGTGCAAGTGAAGCTGAACGGCACCGAGTACTTCAGGAAGTCCATATCGGGCGCCAAGCAGCTCGACGCGCAGGTACTTTATCTAGGCGGGCCCCCTCCCGCCCCGGAGCCCCCGGAAGCGGCCGCGCTAATCGCGTCGCCCCCCTCTCCGTCTCCACCCCCCGCCCCGGCCTCCTCGTCCAGCTCGCCGCCCGTGAACCCGCCGCCCGCGCCGCCGGCCCCCTCCACACCCCCGCCCCCGCCCCCGCCGGTCCCCACGTCCGTGCTGACGGCCCTCACCTCCACGCCGCTCCCCACCGCACCCCCCGAGGAGGAACCCGATGATTCAGCTTACTTCAAAGGCGTCATTCAGGACGTTCAGGTATCGAACGGAGTCAACGTGACCATAGTGGAGTTCTTCCCCCTGCGAGTCCCGGGGCTGGAGCTGCCTCCTCCCTTCGGAGAAGTGTTCCTGCCCCCCGGAGGGGTGCTGCGCGGGGTGGTCTCCGACGACGTGTGCGCCTCGCAGCCCTGCAAGCACAACGCCACCTGCACCAACACCTGGAACGACTTCACGTGCACGTGTCCGCGCGGGTACAAGGGGAAGCAGTGCGGCGACGTGGAGTTCTGCCAGCTGCAGGGCTGCCCCCCCAACTCGCACTGCAGGGACATCGACCAGGGACTGGAGTGCGTGTCCAACGCGACGTTCGACGGCGCCAACACGACGCTGACGTACCGCCTGCGCGACGCGCGCCCCGACCCCCTCGCGCGGCACCCCCTCCCGCCCCCCACCGACCTCACCATCACCTACAGGACAAAGGTAGGGGGCACATTACTGCGAGCGGAGAGCGACCCTTCGGAGGGCCAGGAGGGCTCGTGGTTCGCAGTGGGTGCGTACAAGGAGCGGGTGTCGGTGCAGTGGCGCCTGGACCCCCTGGGCGGTCCCCCCGCCGACCTGCCGCGGGCGTTGCGCATGACGGCGCCGCACTCGCACCTGCACTGGACCACCGTCAAGTTCACCTTCGACAGAGATCAGATCATAGGGTCCTTCGTGGAGGCGGACGGCGAGGAGCGCGTGGGGCTGCGCGGACACATCGACGTGGCGGCGTGGCAGCGCCTGGTGTCCGCGGGCCGCATCCTGCTGGGCGGGGGGGGCCGCGCGCCCGCCGCGCCCCCGCCCCCCACCCTCAACATGGAGACGTCGACCGATCAGGCCTACGAGTTCACGGAGGGCGAGGAGTTTGCTGCCGATAATATAGAAGGGAAATACTTCAAGGGCTGCGTGGGCGCCGTGCACGTGGGCGGGCTGCTGCTGCCGTTCTTCACGGAGGAGAAGCTGTTCGTGGGCCCGGCCGCCGCGCTGCTGGCCGCGCAGCCGCACTACGCGCTCATTTCCGGCGTGCCGTGGGGCGCGGCGGAGGGCGTGGGCTGCGTGCTGTGCCTGGAGGCGGACTGCAACAACGGCGGGCGCTGCGCCGACGTGCGCTCCTCCTACTCCTGCTCCTGTCCGCCCGGGTACTCCGGAGACTACTGCCAGGTGGACATCGACGAGTGCGCCGTGCACGAGTGCCAGCACGGCGCCACGTGCCGCGACCTGGTGGCCGCCTACCGCTGCGAGTGCCCGCCCGGCTTCGAGGGGCAGCTGTGCGAGAGCGACATCGACGAGTGCACGTCGTCCCCGTGCGCCAACGGCGGCACGTGCGTGGACGCGCCCGGCGGCTACACGTGCACGTGCTCGCCGCAGTGGCGCGGGGACACGTGTCGCGAGCCACGCGCCCGCACATGCGCACACTCCCCCTGCGCCCCCGGCGCCGCGCGCTGCACGGACGACCCCCCCGATCCTCCGACCGGGAACAAGTTCACGTGCGAGTGCCGGGAGGGCTACGAGGGCGTGTACTGCGAGAAGCCGTTCTGCGAGGTGACGCCGTGCGTGCACGGGGAGTGCCGCGCCGACGCGCAGGGCGTGCAGTGCGCGTGCGCGCCGGGCTGGACGGGCGCGCGCTGCTCGGAGGAGCGCGACGAGTGCGTGGGCCACTGCCAGCACGGCGGCCGCTGCTACCGGGACTCCGAGCACTTCCTCTGCGACTGCACCGCCACCGGCTGGACGGGCGCGCGCTGCGAGGTGGACGTGGACGAGTGTGCCGAGGGACTGGTCAGCTGCGGGCCCGGGGACTGCCGCAACCTCAACGGGTCCTACACCTGCGTGTGCGGGCGCGGCTACTGCGGCGCGGAGTGCGCGCTCGTGGACCCGTGCGGCGCCGAGCCGTGCCAGCACGGGGCCGTGTGCGAGCAGCGCTGCGCCCGCGAGCCCGACTACGTGTGCCGCTGCACCGACGGCTGGGCCGGCAAGAACTGCACGCAGCAGGCCGTGGCGGAGGGGTCGGAGGGGACCAGCGACAACTCGTCGGCCGTGCTGCTGGGGGTGGGGGGCGCGCTGGTGGCGCTGGCGCTGGCGGGCGGCGCGCTGGGAGCCCTGGGGGCGCAGGCCCGCCGCAAGCGAGCCACTCGCGGCACCTACTCTCCCTCCGGACAGGAGTACTGCAACCCTCGCGCGGAGATGCACCACCACGCGCTCAAGCCGCCTCCCGAAGAGAGACTCATATAG
Protein
MAHVKLVFMFLFVKGRYCETDVDECAMKPKICNNGVCVNTPGSYQCYCRPGYTGDSCEQDIDECLSSPCKNSGTCQNLENNYECVCMEGFEGKDCSVNIDECEASPCAAGSTCVDGVASYSCQCQEGLTGPRCEVDIDDCESQPCQHGGRCVDALNGYSCECGGTGFTGDDCSVNIDECAPAPCRHGATCVDELNDYRCSCHPGFTGKNCEVDINECEGEPCKFGGVCLERSNASLYLAPDAPPVLVGPHDAMLLPPAFYQPFAIDDASGFECVCVPGTAGARCELNIDECASSPCGHGKCVDGVGSYSCDCLPGYEGDHCEIEIDECQRYQPCAHGRCVDQRATYHCSCSAGWGGRNCSVVLQGCAAQPCRNQGSCHPWLRDETEHRFNCSCANGFYGTTCEKITTMSLERSSYAEVNTSREEGYDISFRFKTTLGDGLLAMGIGLTYFFLELSNGRLNLHSSLLNKWEGVFIGSNLNDSNWQKVFVTINSSHLVLAANEEQTIYPISQIEAYNSSVTSFPTTRLGRAGSSFATLTHGPDFFVGCFQDVVVNGQWVLPESGESEGEGPVWRLAGVTAACPRVPQCQPSPCRSGGRCHDHWTSFLCTCPRPHLGDTCQYNYTAATFGHEAARRGSAVRVLVGEAARRAVLAALDISMFIRTRKPTGQIFYLGSLPRYGQTEETQVGASLSNGELLVHLRFNQTPENYTVGGTRLDNGHLHLIEVVRNSTLVQVKLNGTEYFRKSISGAKQLDAQVLYLGGPPPAPEPPEAAALIASPPSPSPPPAPASSSSSPPVNPPPAPPAPSTPPPPPPPVPTSVLTALTSTPLPTAPPEEEPDDSAYFKGVIQDVQVSNGVNVTIVEFFPLRVPGLELPPPFGEVFLPPGGVLRGVVSDDVCASQPCKHNATCTNTWNDFTCTCPRGYKGKQCGDVEFCQLQGCPPNSHCRDIDQGLECVSNATFDGANTTLTYRLRDARPDPLARHPLPPPTDLTITYRTKVGGTLLRAESDPSEGQEGSWFAVGAYKERVSVQWRLDPLGGPPADLPRALRMTAPHSHLHWTTVKFTFDRDQIIGSFVEADGEERVGLRGHIDVAAWQRLVSAGRILLGGGGRAPAAPPPPTLNMETSTDQAYEFTEGEEFAADNIEGKYFKGCVGAVHVGGLLLPFFTEEKLFVGPAAALLAAQPHYALISGVPWGAAEGVGCVLCLEADCNNGGRCADVRSSYSCSCPPGYSGDYCQVDIDECAVHECQHGATCRDLVAAYRCECPPGFEGQLCESDIDECTSSPCANGGTCVDAPGGYTCTCSPQWRGDTCREPRARTCAHSPCAPGAARCTDDPPDPPTGNKFTCECREGYEGVYCEKPFCEVTPCVHGECRADAQGVQCACAPGWTGARCSEERDECVGHCQHGGRCYRDSEHFLCDCTATGWTGARCEVDVDECAEGLVSCGPGDCRNLNGSYTCVCGRGYCGAECALVDPCGAEPCQHGAVCEQRCAREPDYVCRCTDGWAGKNCTQQAVAEGSEGTSDNSSAVLLGVGGALVALALAGGALGALGAQARRKRATRGTYSPSGQEYCNPRAEMHHHALKPPPEERLI
Summary
Uniprot
ProteinModelPortal
PDB
4XBM
E-value=2.09082e-47,
Score=483
Ontologies
GO
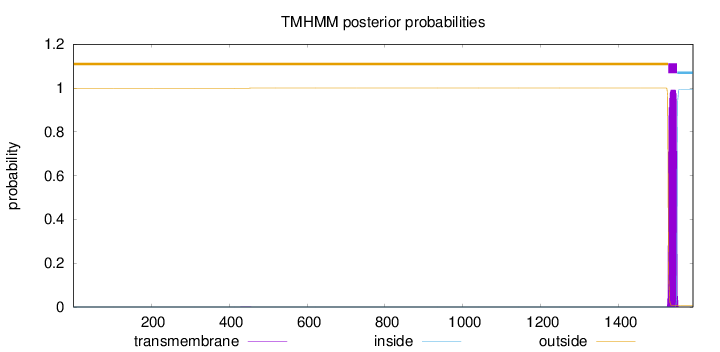
Topology
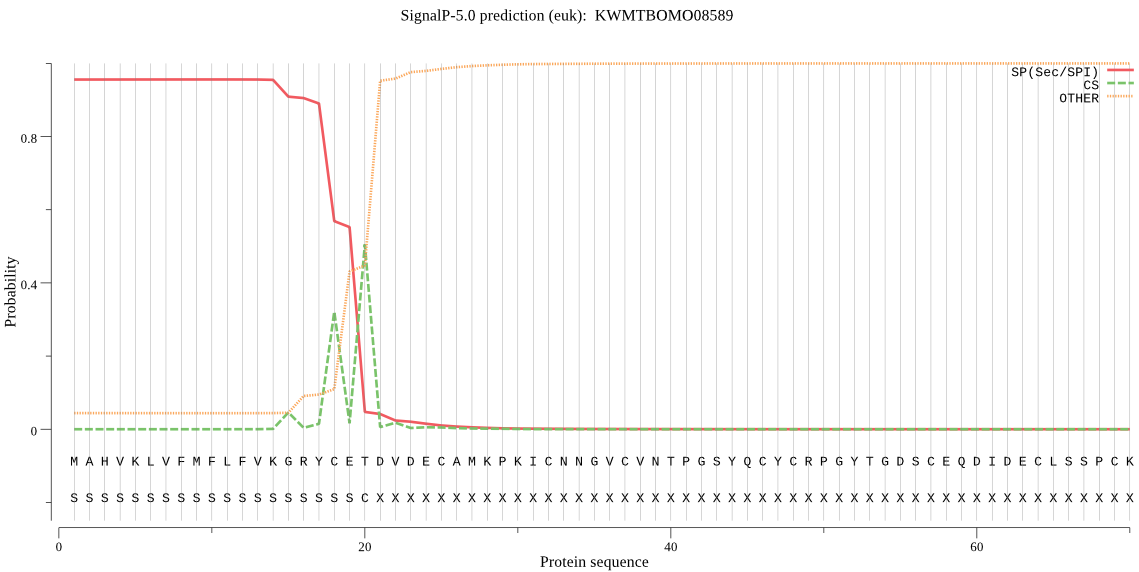
SignalP
Position: 1 - 20,
Likelihood: 0.956273
Length:
1590
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.60532
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00057
outside
1 - 1526
TMhelix
1527 - 1549
inside
1550 - 1590
Population Genetic Test Statistics
Pi
260.111641
Theta
186.204697
Tajima's D
1.596373
CLR
0.824809
CSRT
0.811459427028649
Interpretation
Uncertain
