Pre Gene Modal
BGIBMGA007977
Annotation
PREDICTED:_protein_FAM188A_[Papilio_polytes]
Full name
Ubiquitin carboxyl-terminal hydrolase MINDY-3 homolog
Alternative Name
Deubiquitinating enzyme MINDY-3
Protein CARP homolog
Protein CARP homolog
Location in the cell
Cytoplasmic Reliability : 1.35 PlasmaMembrane Reliability : 1.415
Sequence
CDS
ATGAGTGCGAGTAGCGTGGCGCTGGAGCGCGAGCTGGCAGGCACGCGACGCTTGCTGTGGGGGGACCACGTCAAAGAAGATGTTTTCCGACGTTGGGCGCAAGGTTTTCAATTCAGTCCTGATGAACCCAGCGCCCTCATTCAGCAGGAGGGTGGACCTTGCGCTGCGATCGCCCCAGTACAAGGATTTTTGCTTAAGATTCTCCTATCAGAGACTCCAGGACATAGTTTTCAAGACTTAACATCGGAGAAGTGCAACTCTTTACTTGTGAGAGCTGTGTGTACGATTTTAAGCCAGTGTCTAGCACCTAAATACAATGTAGCTGTTTACCGGAAGAATGAGTCTGGTGCGGAAACAAGTGGAAGTAACAGCGTTAGTGTGGATGAACACTCTAGTGATACCATTGAACATTTCCATCGAAGAATCCAGGTACATTCGTTTCAAAATCTGTCAGAAGTGGAGGCATTTTACACAAGGAACATTAGGATATTGAAGGACAAATATGGTGTTCTACTTCTGTTATATAGTGTGATTCTCAGTAAGGGTATAAGTACAGTAGAAGCAGAGCTCTCTGAACTCTCAGACCCCCTGATACATTCCACATATGGGTATGGATCACAAGGTCTCATCAATTTGATGCTGACTGGACGAGCAGTGGCACATGTGTGGGATCATGATCAAGTTGTCAGTGGTTTAAGACTTAGAGGCATAGAAAAACAAAATGACATTGGTTTCCTGACTATCATGGAGCATATGCAATACTGCACTGTAGGGTCGTTTTACAAGAGCCCTAAACATCCTGTTTGGGTACTGGCTTCCGAGACTCACCTGACTGTGCTTTTCTCTCTAGAGCGTCGATTGGCAGCACCAGAGAGTGCAGGAGAATCTGCTGAACGAATATTCAGATCATTTGATCCTGAAGGCAACAATTTCATTCCTTCAGTAGCTTTGCAGGATGTTCTGTGCGCTGCAGACTTAGTCAGTGAACCAGAATATGTGGAATTAATGAGGCGGAAATTAGACACTGAAAACTTGGGAATAATTTTGTTAAGTACTTTCATGGATGAATTTTTCCCAGGATGTGAACGTGGTGCTCCTGACACATTCACCCTTCACCATTACAATGGTCTTGAGCGTAGTAACCCAGGTGGCAGAGTTGTGTACCGCACTGGTCGAGCAGCTTTGCTCGAGTGCCCGATGCGTGCTGCCACCACAGACCCCATGTTGACCTGTCTGCAGACTAAGTGGCCCAGCATTGATGTTGTCTGGGACGATGGACATTCACCCTCGCTCAATTAA
Protein
MSASSVALERELAGTRRLLWGDHVKEDVFRRWAQGFQFSPDEPSALIQQEGGPCAAIAPVQGFLLKILLSETPGHSFQDLTSEKCNSLLVRAVCTILSQCLAPKYNVAVYRKNESGAETSGSNSVSVDEHSSDTIEHFHRRIQVHSFQNLSEVEAFYTRNIRILKDKYGVLLLLYSVILSKGISTVEAELSELSDPLIHSTYGYGSQGLINLMLTGRAVAHVWDHDQVVSGLRLRGIEKQNDIGFLTIMEHMQYCTVGSFYKSPKHPVWVLASETHLTVLFSLERRLAAPESAGESAERIFRSFDPEGNNFIPSVALQDVLCAADLVSEPEYVELMRRKLDTENLGIILLSTFMDEFFPGCERGAPDTFTLHHYNGLERSNPGGRVVYRTGRAALLECPMRAATTDPMLTCLQTKWPSIDVVWDDGHSPSLN
Summary
Description
Hydrolase that can remove 'Lys-48'-linked conjugated ubiquitin from proteins.
Catalytic Activity
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
Similarity
Belongs to the MINDY deubiquitinase family. FAM188 subfamily.
Keywords
Complete proteome
Hydrolase
Phosphoprotein
Protease
Reference proteome
Thiol protease
Ubl conjugation pathway
Feature
chain Ubiquitin carboxyl-terminal hydrolase MINDY-3 homolog
Uniprot
H9JEN0
A0A3S2L923
A0A0N1IQ86
A0A194PPS2
A0A2W1BD68
A0A2A4JG99
+ More
A0A2H1VV89 S4Q0A4 A0A0L7LE07 A0A0K8TKV3 A0A1A9X4J8 A0A1A9UU33 A0A1B0FK59 A0A1A9ZB82 A0A1A9XKB8 A0A1B0ALY5 A0A067R0Q9 W8AT25 A0A0K8UNR8 A0A0K8TWG7 A0A034W2B7 A0A2J7QUX6 A0A1I8MBV1 A0A0A1XJA2 A0A0A1XRV6 A0A1I8P9S4 A0A195CWK4 A0A1L8DAV1 E2BWV3 B4L3A7 A0A0Q9WMI6 F4WZI4 A0A158NH15 Q16VR7 A0A195DQ81 A0A195ETZ9 A0A195BXV1 W5JN04 E9IE64 Q7QFC9 A0A182YCS4 A0A026VZY2 B4M3T3 A0A182FMN6 A0A182RKY3 X2JL49 Q9VWN5 D2A5F4 A0A1B6M376 A0A2M3ZI78 A0A182KGW2 A0A2M4BJM0 E2AGX5 A0A2M4BJQ1 B4I6Z0 B4PWH1 A0A1W4U9Y0 B3NUQ4 A0A0L0C0L0 A0A182QYT7 A0A0J7NI26 A0A1B6G384 A0A1B6HXE0 A0A182P181 A0A023ETI2 T1HC57 A0A0N7Z936 A0A182MVQ0 A0A1Y1NHQ8 A0A069DU89 A0A0L7QN75 A0A023F2W2 A0A224XCX8 B0XCH7 A0A182W5T3 A0A0T6BGQ4 A0A088AEQ0 K7IT19 A0A1B6DPN6 A0A2A3ECX4 A0A1Q3F7X3 A0A232F6D5 A0A1Q3F7R6 A0A1Q3F7U5 A0A0N8NZN7 A0A182HAP8 N6TTA5 A0A3L8DFF7 B7S8F1 A0A2M3ZZZ6 A0A2M4A039 A0A146LSQ5 A0A0A9YG83 A0A0K8S6T2 A0A1J1J3Y8 T1IVP6 U4UXQ6 A0A182U586 A0A1S3CWF9
A0A2H1VV89 S4Q0A4 A0A0L7LE07 A0A0K8TKV3 A0A1A9X4J8 A0A1A9UU33 A0A1B0FK59 A0A1A9ZB82 A0A1A9XKB8 A0A1B0ALY5 A0A067R0Q9 W8AT25 A0A0K8UNR8 A0A0K8TWG7 A0A034W2B7 A0A2J7QUX6 A0A1I8MBV1 A0A0A1XJA2 A0A0A1XRV6 A0A1I8P9S4 A0A195CWK4 A0A1L8DAV1 E2BWV3 B4L3A7 A0A0Q9WMI6 F4WZI4 A0A158NH15 Q16VR7 A0A195DQ81 A0A195ETZ9 A0A195BXV1 W5JN04 E9IE64 Q7QFC9 A0A182YCS4 A0A026VZY2 B4M3T3 A0A182FMN6 A0A182RKY3 X2JL49 Q9VWN5 D2A5F4 A0A1B6M376 A0A2M3ZI78 A0A182KGW2 A0A2M4BJM0 E2AGX5 A0A2M4BJQ1 B4I6Z0 B4PWH1 A0A1W4U9Y0 B3NUQ4 A0A0L0C0L0 A0A182QYT7 A0A0J7NI26 A0A1B6G384 A0A1B6HXE0 A0A182P181 A0A023ETI2 T1HC57 A0A0N7Z936 A0A182MVQ0 A0A1Y1NHQ8 A0A069DU89 A0A0L7QN75 A0A023F2W2 A0A224XCX8 B0XCH7 A0A182W5T3 A0A0T6BGQ4 A0A088AEQ0 K7IT19 A0A1B6DPN6 A0A2A3ECX4 A0A1Q3F7X3 A0A232F6D5 A0A1Q3F7R6 A0A1Q3F7U5 A0A0N8NZN7 A0A182HAP8 N6TTA5 A0A3L8DFF7 B7S8F1 A0A2M3ZZZ6 A0A2M4A039 A0A146LSQ5 A0A0A9YG83 A0A0K8S6T2 A0A1J1J3Y8 T1IVP6 U4UXQ6 A0A182U586 A0A1S3CWF9
EC Number
3.4.19.12
Pubmed
19121390
26354079
28756777
23622113
26227816
26369729
+ More
24845553 24495485 25348373 25315136 25830018 20798317 17994087 21719571 21347285 17510324 20920257 23761445 21282665 12364791 14747013 17210077 25244985 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 18327897 18362917 19820115 17550304 26108605 24945155 27129103 28004739 26334808 25474469 20075255 28648823 26483478 23537049 30249741 26823975 25401762
24845553 24495485 25348373 25315136 25830018 20798317 17994087 21719571 21347285 17510324 20920257 23761445 21282665 12364791 14747013 17210077 25244985 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 18327897 18362917 19820115 17550304 26108605 24945155 27129103 28004739 26334808 25474469 20075255 28648823 26483478 23537049 30249741 26823975 25401762
EMBL
BABH01003338
RSAL01000083
RVE48438.1
KQ459717
KPJ20313.1
KQ459596
+ More
KPI95312.1 KZ150445 PZC70850.1 NWSH01001686 PCG70422.1 ODYU01004604 SOQ44666.1 GAIX01000133 JAA92427.1 JTDY01001529 KOB73635.1 GDAI01002837 JAI14766.1 CCAG010004878 JXJN01000215 KK853123 KDR11065.1 GAMC01014595 JAB91960.1 GDHF01031509 GDHF01023985 GDHF01015813 JAI20805.1 JAI28329.1 JAI36501.1 GDHF01033683 JAI18631.1 GAKP01010118 JAC48834.1 NEVH01010480 PNF32390.1 GBXI01003322 JAD10970.1 GBXI01000561 JAD13731.1 KQ977259 KYN04534.1 GFDF01010495 JAV03589.1 GL451188 EFN79817.1 CH933810 EDW07035.1 KRF93917.1 GL888479 EGI60428.1 ADTU01001620 CH477583 EAT38655.1 KQ980612 KYN15080.1 KQ981979 KYN31382.1 KQ976398 KYM92771.1 ADMH02001081 ETN64154.1 GL762556 EFZ21187.1 AAAB01008846 EAA06126.5 KK107519 EZA49363.1 CH940651 EDW65458.2 AE014298 AHN59887.1 AY058544 KQ971345 EFA05086.2 GEBQ01009601 JAT30376.1 GGFM01007387 MBW28138.1 GGFJ01004106 MBW53247.1 GL439433 EFN67311.1 GGFJ01004093 MBW53234.1 CH480823 EDW56088.1 CM000162 EDX02789.1 CH954180 EDV46651.1 JRES01001065 KNC25817.1 AXCN02002227 LBMM01004724 KMQ92140.1 GECZ01012897 JAS56872.1 GECU01028367 JAS79339.1 GAPW01000980 JAC12618.1 ACPB03010146 GDKW01001643 JAI54952.1 AXCM01006171 GEZM01001969 JAV97433.1 GBGD01001553 JAC87336.1 KQ414874 KOC59931.1 GBBI01003266 JAC15446.1 GFTR01006151 JAW10275.1 DS232696 EDS44839.1 LJIG01000404 KRT86536.1 AAZX01005110 GEDC01009652 JAS27646.1 KZ288301 PBC28871.1 GFDL01011380 JAV23665.1 NNAY01000889 OXU26050.1 GFDL01011431 JAV23614.1 GFDL01011401 JAV23644.1 CH902635 KPU74836.1 JXUM01123047 KQ566673 KXJ69917.1 APGK01052864 KB741216 ENN72495.1 QOIP01000009 RLU18628.1 EF710645 ACE75176.1 GGFK01000750 MBW34071.1 GGFK01000865 MBW34186.1 GDHC01007821 JAQ10808.1 GBHO01011529 GBHO01011527 GBHO01011524 JAG32075.1 JAG32077.1 JAG32080.1 GBRD01017382 JAG48445.1 CVRI01000070 CRL07128.1 JH431589 KB632404 ERL95075.1
KPI95312.1 KZ150445 PZC70850.1 NWSH01001686 PCG70422.1 ODYU01004604 SOQ44666.1 GAIX01000133 JAA92427.1 JTDY01001529 KOB73635.1 GDAI01002837 JAI14766.1 CCAG010004878 JXJN01000215 KK853123 KDR11065.1 GAMC01014595 JAB91960.1 GDHF01031509 GDHF01023985 GDHF01015813 JAI20805.1 JAI28329.1 JAI36501.1 GDHF01033683 JAI18631.1 GAKP01010118 JAC48834.1 NEVH01010480 PNF32390.1 GBXI01003322 JAD10970.1 GBXI01000561 JAD13731.1 KQ977259 KYN04534.1 GFDF01010495 JAV03589.1 GL451188 EFN79817.1 CH933810 EDW07035.1 KRF93917.1 GL888479 EGI60428.1 ADTU01001620 CH477583 EAT38655.1 KQ980612 KYN15080.1 KQ981979 KYN31382.1 KQ976398 KYM92771.1 ADMH02001081 ETN64154.1 GL762556 EFZ21187.1 AAAB01008846 EAA06126.5 KK107519 EZA49363.1 CH940651 EDW65458.2 AE014298 AHN59887.1 AY058544 KQ971345 EFA05086.2 GEBQ01009601 JAT30376.1 GGFM01007387 MBW28138.1 GGFJ01004106 MBW53247.1 GL439433 EFN67311.1 GGFJ01004093 MBW53234.1 CH480823 EDW56088.1 CM000162 EDX02789.1 CH954180 EDV46651.1 JRES01001065 KNC25817.1 AXCN02002227 LBMM01004724 KMQ92140.1 GECZ01012897 JAS56872.1 GECU01028367 JAS79339.1 GAPW01000980 JAC12618.1 ACPB03010146 GDKW01001643 JAI54952.1 AXCM01006171 GEZM01001969 JAV97433.1 GBGD01001553 JAC87336.1 KQ414874 KOC59931.1 GBBI01003266 JAC15446.1 GFTR01006151 JAW10275.1 DS232696 EDS44839.1 LJIG01000404 KRT86536.1 AAZX01005110 GEDC01009652 JAS27646.1 KZ288301 PBC28871.1 GFDL01011380 JAV23665.1 NNAY01000889 OXU26050.1 GFDL01011431 JAV23614.1 GFDL01011401 JAV23644.1 CH902635 KPU74836.1 JXUM01123047 KQ566673 KXJ69917.1 APGK01052864 KB741216 ENN72495.1 QOIP01000009 RLU18628.1 EF710645 ACE75176.1 GGFK01000750 MBW34071.1 GGFK01000865 MBW34186.1 GDHC01007821 JAQ10808.1 GBHO01011529 GBHO01011527 GBHO01011524 JAG32075.1 JAG32077.1 JAG32080.1 GBRD01017382 JAG48445.1 CVRI01000070 CRL07128.1 JH431589 KB632404 ERL95075.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000218220
UP000037510
+ More
UP000091820 UP000078200 UP000092444 UP000092445 UP000092443 UP000092460 UP000027135 UP000235965 UP000095301 UP000095300 UP000078542 UP000008237 UP000009192 UP000007755 UP000005205 UP000008820 UP000078492 UP000078541 UP000078540 UP000000673 UP000007062 UP000076408 UP000053097 UP000008792 UP000069272 UP000075900 UP000000803 UP000007266 UP000075881 UP000000311 UP000001292 UP000002282 UP000192221 UP000008711 UP000037069 UP000075886 UP000036403 UP000075885 UP000015103 UP000075883 UP000053825 UP000002320 UP000075920 UP000005203 UP000002358 UP000242457 UP000215335 UP000007801 UP000069940 UP000249989 UP000019118 UP000279307 UP000183832 UP000030742 UP000075902 UP000079169
UP000091820 UP000078200 UP000092444 UP000092445 UP000092443 UP000092460 UP000027135 UP000235965 UP000095301 UP000095300 UP000078542 UP000008237 UP000009192 UP000007755 UP000005205 UP000008820 UP000078492 UP000078541 UP000078540 UP000000673 UP000007062 UP000076408 UP000053097 UP000008792 UP000069272 UP000075900 UP000000803 UP000007266 UP000075881 UP000000311 UP000001292 UP000002282 UP000192221 UP000008711 UP000037069 UP000075886 UP000036403 UP000075885 UP000015103 UP000075883 UP000053825 UP000002320 UP000075920 UP000005203 UP000002358 UP000242457 UP000215335 UP000007801 UP000069940 UP000249989 UP000019118 UP000279307 UP000183832 UP000030742 UP000075902 UP000079169
Pfam
Interpro
SUPFAM
SSF47473
SSF47473
ProteinModelPortal
H9JEN0
A0A3S2L923
A0A0N1IQ86
A0A194PPS2
A0A2W1BD68
A0A2A4JG99
+ More
A0A2H1VV89 S4Q0A4 A0A0L7LE07 A0A0K8TKV3 A0A1A9X4J8 A0A1A9UU33 A0A1B0FK59 A0A1A9ZB82 A0A1A9XKB8 A0A1B0ALY5 A0A067R0Q9 W8AT25 A0A0K8UNR8 A0A0K8TWG7 A0A034W2B7 A0A2J7QUX6 A0A1I8MBV1 A0A0A1XJA2 A0A0A1XRV6 A0A1I8P9S4 A0A195CWK4 A0A1L8DAV1 E2BWV3 B4L3A7 A0A0Q9WMI6 F4WZI4 A0A158NH15 Q16VR7 A0A195DQ81 A0A195ETZ9 A0A195BXV1 W5JN04 E9IE64 Q7QFC9 A0A182YCS4 A0A026VZY2 B4M3T3 A0A182FMN6 A0A182RKY3 X2JL49 Q9VWN5 D2A5F4 A0A1B6M376 A0A2M3ZI78 A0A182KGW2 A0A2M4BJM0 E2AGX5 A0A2M4BJQ1 B4I6Z0 B4PWH1 A0A1W4U9Y0 B3NUQ4 A0A0L0C0L0 A0A182QYT7 A0A0J7NI26 A0A1B6G384 A0A1B6HXE0 A0A182P181 A0A023ETI2 T1HC57 A0A0N7Z936 A0A182MVQ0 A0A1Y1NHQ8 A0A069DU89 A0A0L7QN75 A0A023F2W2 A0A224XCX8 B0XCH7 A0A182W5T3 A0A0T6BGQ4 A0A088AEQ0 K7IT19 A0A1B6DPN6 A0A2A3ECX4 A0A1Q3F7X3 A0A232F6D5 A0A1Q3F7R6 A0A1Q3F7U5 A0A0N8NZN7 A0A182HAP8 N6TTA5 A0A3L8DFF7 B7S8F1 A0A2M3ZZZ6 A0A2M4A039 A0A146LSQ5 A0A0A9YG83 A0A0K8S6T2 A0A1J1J3Y8 T1IVP6 U4UXQ6 A0A182U586 A0A1S3CWF9
A0A2H1VV89 S4Q0A4 A0A0L7LE07 A0A0K8TKV3 A0A1A9X4J8 A0A1A9UU33 A0A1B0FK59 A0A1A9ZB82 A0A1A9XKB8 A0A1B0ALY5 A0A067R0Q9 W8AT25 A0A0K8UNR8 A0A0K8TWG7 A0A034W2B7 A0A2J7QUX6 A0A1I8MBV1 A0A0A1XJA2 A0A0A1XRV6 A0A1I8P9S4 A0A195CWK4 A0A1L8DAV1 E2BWV3 B4L3A7 A0A0Q9WMI6 F4WZI4 A0A158NH15 Q16VR7 A0A195DQ81 A0A195ETZ9 A0A195BXV1 W5JN04 E9IE64 Q7QFC9 A0A182YCS4 A0A026VZY2 B4M3T3 A0A182FMN6 A0A182RKY3 X2JL49 Q9VWN5 D2A5F4 A0A1B6M376 A0A2M3ZI78 A0A182KGW2 A0A2M4BJM0 E2AGX5 A0A2M4BJQ1 B4I6Z0 B4PWH1 A0A1W4U9Y0 B3NUQ4 A0A0L0C0L0 A0A182QYT7 A0A0J7NI26 A0A1B6G384 A0A1B6HXE0 A0A182P181 A0A023ETI2 T1HC57 A0A0N7Z936 A0A182MVQ0 A0A1Y1NHQ8 A0A069DU89 A0A0L7QN75 A0A023F2W2 A0A224XCX8 B0XCH7 A0A182W5T3 A0A0T6BGQ4 A0A088AEQ0 K7IT19 A0A1B6DPN6 A0A2A3ECX4 A0A1Q3F7X3 A0A232F6D5 A0A1Q3F7R6 A0A1Q3F7U5 A0A0N8NZN7 A0A182HAP8 N6TTA5 A0A3L8DFF7 B7S8F1 A0A2M3ZZZ6 A0A2M4A039 A0A146LSQ5 A0A0A9YG83 A0A0K8S6T2 A0A1J1J3Y8 T1IVP6 U4UXQ6 A0A182U586 A0A1S3CWF9
Ontologies
PANTHER
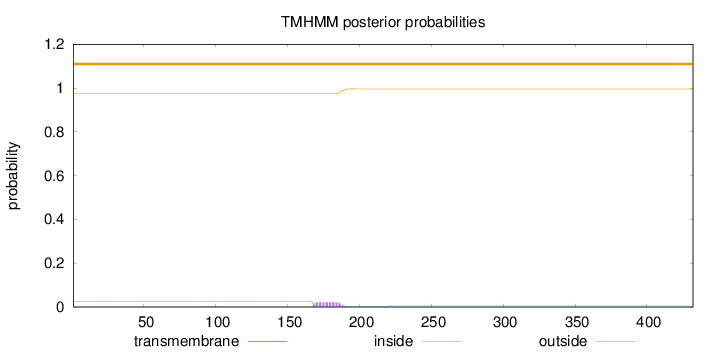
Topology
Length:
432
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.44509
Exp number, first 60 AAs:
0.00045
Total prob of N-in:
0.02451
outside
1 - 432
Population Genetic Test Statistics
Pi
145.783613
Theta
168.555348
Tajima's D
-0.30607
CLR
16.425682
CSRT
0.288335583220839
Interpretation
Uncertain
