Gene
KWMTBOMO08583
Annotation
PREDICTED:_apoptosis_regulatory_protein_Siva-like_[Amyelois_transitella]
Full name
Apoptosis regulatory protein Siva
Alternative Name
CD27-binding protein
Location in the cell
Extracellular Reliability : 2.055 Nuclear Reliability : 2.427
Sequence
CDS
ATGACAAAACGAACGAACCCTTTTATCGATGATTTTCTTCCGCAAAGTAAAATTCATGTTAGTTTAAAAAAGTACGATAAACACAGCACAGACAATGAAAAACGACTGAAGAAAGTTTATGAAAAAACCCTACAAATGTTATTTAATGGCGCCAAAAAGGGTACAACTACTGAAACCAACAACAACTTGCAGGATAGTAAAGTGAGAAAAGACAAAAAAAAACAATTATTTATCGGCAAAGATGGCAATCTACTACATTCTGGTTGTCTGATAGAGACCCAATCAATAAGAAAAAACTGTCATTGCGGTGTTACATCAGAGACCAAGTGTGCATATTGTGAATTTGTACTATGTGATTCATGTCTCCACACCTGCGCTGGTTGTGAGCAGGTTTACTGTAACAAATGCTTGTTTGTAGGGTCAGAAGGTTCTGAAATCTGTGTATCCTGTTATAGTTAA
Protein
MTKRTNPFIDDFLPQSKIHVSLKKYDKHSTDNEKRLKKVYEKTLQMLFNGAKKGTTTETNNNLQDSKVRKDKKKQLFIGKDGNLLHSGCLIETQSIRKNCHCGVTSETKCAYCEFVLCDSCLHTCAGCEQVYCNKCLFVGSEGSEICVSCYS
Summary
Description
Induces CD27-mediated apoptosis. Inhibits BCL2L1 isoform Bcl-x(L) anti-apoptotic activity. Inhibits activation of NF-kappa-B and promotes T-cell receptor-mediated apoptosis (By similarity).
Induces CD27-mediated apoptosis. Inhibits BCL2L1 isoform Bcl-x(L) anti-apoptotic activity. Inhibits activation of NF-kappa-B and promotes T-cell receptor-mediated apoptosis.
Induces CD27-mediated apoptosis. Inhibits BCL2L1 isoform Bcl-x(L) anti-apoptotic activity. Inhibits activation of NF-kappa-B and promotes T-cell receptor-mediated apoptosis.
Cofactor
Zn(2+)
Subunit
Binds through its N-terminal region to the C-terminus of CD27 and to PXMP2/PMP22. Binds to the C-terminus of TNFRSF18/GITR. Isoform 1 binds to BCL2L1/BCLX isoform Bcl-x(L) but not to BAX.
Binds through its N-terminal region to the C-terminus of CD27 and to PXMP2/PMP22. Binds to the C-terminus of TNFRSF18/GITR. Binds to BCL2L1/BCLX isoform Bcl-x(L) but not to BAX (By similarity).
(Microbial infection) Interacts with coxsackievirus B3 capsid protein VP2; this interaction inhibits the binding of SIVA1 to CD27.
Binds through its N-terminal region to the C-terminus of CD27 and to PXMP2/PMP22. Binds to the C-terminus of TNFRSF18/GITR. Binds to BCL2L1/BCLX isoform Bcl-x(L) but not to BAX (By similarity).
(Microbial infection) Interacts with coxsackievirus B3 capsid protein VP2; this interaction inhibits the binding of SIVA1 to CD27.
Keywords
Alternative splicing
Apoptosis
Complete proteome
Cytoplasm
Metal-binding
Nucleus
Phosphoprotein
Reference proteome
Zinc
Host-virus interaction
Pharmaceutical
Feature
chain Apoptosis regulatory protein Siva
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A3S2NTU8
A0A2H1VTR3
A0A2W1B8P2
A0A2A4JWK7
A0A0N1IK27
A0A194PPD8
+ More
S4NMZ4 A0A310SII5 A0A423T5T0 A0A0C9R2L2 A0A3Q8CBS8 O54926 Q3TFC3 A0A088ARN6 A0A026WT68 A0A286Y1S5 A0A2A3ELS2 A0A3Q2YAG8 A0A1E1X0W2 A0A2Y9T932 A0A340X8V0 A0A0N0U4X4 A4FTX4 A0A2U4B8G8 A0A3B3VE50 A0A444SCK9 A0A2U3V608 A0A0L7RDK0 I3MN94 A0A154PA52 A0A2R5LG96 P59692 A0A091CUX7 A0A3B5KGM8 A0A250YB36 A0A341CPU3 Q4RLN8 E9I8P0 A0A3Q3LDP7 V5HE68 G3X7B3 A0A452FIV7 A0A3Q7QA12 A0A1B0GLE0 A0A087XWY1 A0A3B3WND7 A0A3P9QBY1 A0A3B3RIN6 A0A2Y9NJS2 A0A2U3VL24 A0A2U3Y3U7 A0A3B5LRK6 A0A1B6C316 A0A1U7TIP2 A0A1S2ZF59 M3ZWX5 K9IWK9 A0A060WLI1 G5AWI6 A0A1B0CA90 A0A158NHF5 A0A315UYW8 M3VK34 A0A2R8MJ08 A0A2Y9G8P6 G3V439 A0A452IW01 A0A3P8UHT9 B7ZAZ2 B4DTY2 D2I109 H2MR92 A0A452QY36 A0A3Q7WD01 G1LMR9 G3V3U1 A0A2I2YVG9 F7HYP0 A0A3P9LR78 F7E257 J9JMK6 G3RB25 H2Q902 O15304 A0A1U7QL07 U3AQ02 A0A195D4F3 A0A3Q7U526 C4WUU7 A0A1A8HLF8 A0A151XFC9 A0A3Q7TTI3 A0A2K6S859 A0A3Q7TFG3 A0A2J8QLA6 A0A2J8QL97 A0A2J8SZ82 A0A2J8SZ79 A0A2J8QLI8 A0A1A8JQS9
S4NMZ4 A0A310SII5 A0A423T5T0 A0A0C9R2L2 A0A3Q8CBS8 O54926 Q3TFC3 A0A088ARN6 A0A026WT68 A0A286Y1S5 A0A2A3ELS2 A0A3Q2YAG8 A0A1E1X0W2 A0A2Y9T932 A0A340X8V0 A0A0N0U4X4 A4FTX4 A0A2U4B8G8 A0A3B3VE50 A0A444SCK9 A0A2U3V608 A0A0L7RDK0 I3MN94 A0A154PA52 A0A2R5LG96 P59692 A0A091CUX7 A0A3B5KGM8 A0A250YB36 A0A341CPU3 Q4RLN8 E9I8P0 A0A3Q3LDP7 V5HE68 G3X7B3 A0A452FIV7 A0A3Q7QA12 A0A1B0GLE0 A0A087XWY1 A0A3B3WND7 A0A3P9QBY1 A0A3B3RIN6 A0A2Y9NJS2 A0A2U3VL24 A0A2U3Y3U7 A0A3B5LRK6 A0A1B6C316 A0A1U7TIP2 A0A1S2ZF59 M3ZWX5 K9IWK9 A0A060WLI1 G5AWI6 A0A1B0CA90 A0A158NHF5 A0A315UYW8 M3VK34 A0A2R8MJ08 A0A2Y9G8P6 G3V439 A0A452IW01 A0A3P8UHT9 B7ZAZ2 B4DTY2 D2I109 H2MR92 A0A452QY36 A0A3Q7WD01 G1LMR9 G3V3U1 A0A2I2YVG9 F7HYP0 A0A3P9LR78 F7E257 J9JMK6 G3RB25 H2Q902 O15304 A0A1U7QL07 U3AQ02 A0A195D4F3 A0A3Q7U526 C4WUU7 A0A1A8HLF8 A0A151XFC9 A0A3Q7TTI3 A0A2K6S859 A0A3Q7TFG3 A0A2J8QLA6 A0A2J8QL97 A0A2J8SZ82 A0A2J8SZ79 A0A2J8QLI8 A0A1A8JQS9
Pubmed
28756777
26354079
23622113
10597319
16141072
15489334
+ More
9177220 10756043 12478477 15105421 10349636 11042159 11076861 11217851 12466851 16141073 24508170 21993624 28503490 15057822 9853261 21551351 28087693 15496914 21282665 25765539 19393038 29240929 23542700 24755649 21993625 21347285 29703783 12508121 24275569 28562605 24487278 20010809 17554307 22398555 25243066 19892987 16136131 11278261 11601913 12011449 14739602 15034012 15958577 16683188 16491128
9177220 10756043 12478477 15105421 10349636 11042159 11076861 11217851 12466851 16141073 24508170 21993624 28503490 15057822 9853261 21551351 28087693 15496914 21282665 25765539 19393038 29240929 23542700 24755649 21993625 21347285 29703783 12508121 24275569 28562605 24487278 20010809 17554307 22398555 25243066 19892987 16136131 11278261 11601913 12011449 14739602 15034012 15958577 16683188 16491128
EMBL
RSAL01000083
RVE48433.1
ODYU01004391
SOQ44219.1
KZ150445
PZC70845.1
+ More
NWSH01000539 PCG75780.1 KQ459717 KPJ20318.1 KQ459596 KPI95306.1 GAIX01012439 JAA80121.1 KQ760869 OAD58766.1 QCYY01002239 ROT71847.1 GBYB01007077 JAG76844.1 KY056151 ATX63073.1 AF033115 AF033113 AF033112 AF033114 AK010562 AK017570 AK153115 BC012279 AK169200 BAE40975.1 KK107109 EZA59207.1 AAKN02051999 KZ288222 PBC32109.1 GFAC01006314 JAT92874.1 KQ435814 KOX72674.1 AABR07073265 AC141959 BC088866 AAH88866.1 SAUD01042300 RXG51111.1 KQ414614 KOC68860.1 AGTP01078354 KQ434856 KZC08732.1 GGLE01004415 MBY08541.1 KN124093 KFO22302.1 GFFW01003978 JAV40810.1 CAAE01015019 CAG10694.1 GL761591 EFZ23069.1 GANP01011001 JAB73467.1 LWLT01000019 AJWK01034780 AYCK01003396 GEDC01029703 JAS07595.1 GABZ01008224 JAA45301.1 FR904523 CDQ65949.1 JH167269 GEBF01000643 EHB01397.1 JAO02990.1 AJWK01003422 ADTU01015734 NHOQ01002481 PWA16605.1 GACC01000340 JAA74244.1 AL583722 AK316457 BAH14828.1 AK300415 AK316550 BAG62144.1 BAG65628.1 GL193937 EFB21661.1 ACTA01012284 ACTA01020284 CABD030094324 GAMP01007779 JAB44976.1 ABLF02022108 AACZ04057246 GABF01002560 GABD01000115 JAA19585.1 JAA32985.1 U82938 AF033111 BC034562 AF401214 BK000018 GAMT01009688 GAMS01008581 GAMQ01006549 JAB02173.1 JAB14555.1 JAB35302.1 KQ976870 KYN07780.1 AK341228 BAH71667.1 HAEB01007997 HAEC01016050 SBQ84271.1 KQ982194 KYQ59094.1 NBAG03000028 PNI97066.1 PNI97065.1 NDHI03003534 PNJ26075.1 PNJ26076.1 PNI97061.1 HAED01017306 HAEE01002247 SBR22267.1
NWSH01000539 PCG75780.1 KQ459717 KPJ20318.1 KQ459596 KPI95306.1 GAIX01012439 JAA80121.1 KQ760869 OAD58766.1 QCYY01002239 ROT71847.1 GBYB01007077 JAG76844.1 KY056151 ATX63073.1 AF033115 AF033113 AF033112 AF033114 AK010562 AK017570 AK153115 BC012279 AK169200 BAE40975.1 KK107109 EZA59207.1 AAKN02051999 KZ288222 PBC32109.1 GFAC01006314 JAT92874.1 KQ435814 KOX72674.1 AABR07073265 AC141959 BC088866 AAH88866.1 SAUD01042300 RXG51111.1 KQ414614 KOC68860.1 AGTP01078354 KQ434856 KZC08732.1 GGLE01004415 MBY08541.1 KN124093 KFO22302.1 GFFW01003978 JAV40810.1 CAAE01015019 CAG10694.1 GL761591 EFZ23069.1 GANP01011001 JAB73467.1 LWLT01000019 AJWK01034780 AYCK01003396 GEDC01029703 JAS07595.1 GABZ01008224 JAA45301.1 FR904523 CDQ65949.1 JH167269 GEBF01000643 EHB01397.1 JAO02990.1 AJWK01003422 ADTU01015734 NHOQ01002481 PWA16605.1 GACC01000340 JAA74244.1 AL583722 AK316457 BAH14828.1 AK300415 AK316550 BAG62144.1 BAG65628.1 GL193937 EFB21661.1 ACTA01012284 ACTA01020284 CABD030094324 GAMP01007779 JAB44976.1 ABLF02022108 AACZ04057246 GABF01002560 GABD01000115 JAA19585.1 JAA32985.1 U82938 AF033111 BC034562 AF401214 BK000018 GAMT01009688 GAMS01008581 GAMQ01006549 JAB02173.1 JAB14555.1 JAB35302.1 KQ976870 KYN07780.1 AK341228 BAH71667.1 HAEB01007997 HAEC01016050 SBQ84271.1 KQ982194 KYQ59094.1 NBAG03000028 PNI97066.1 PNI97065.1 NDHI03003534 PNJ26075.1 PNJ26076.1 PNI97061.1 HAED01017306 HAEE01002247 SBR22267.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000283509
UP000000589
+ More
UP000005203 UP000053097 UP000005447 UP000242457 UP000264820 UP000248484 UP000265300 UP000053105 UP000002494 UP000245320 UP000261500 UP000288706 UP000053825 UP000005215 UP000076502 UP000028990 UP000005226 UP000252040 UP000007303 UP000261640 UP000009136 UP000291000 UP000286641 UP000092461 UP000028760 UP000261480 UP000242638 UP000261540 UP000248483 UP000245340 UP000245341 UP000261380 UP000189704 UP000079721 UP000002852 UP000193380 UP000006813 UP000005205 UP000008227 UP000008225 UP000248481 UP000005640 UP000291020 UP000265120 UP000001038 UP000291022 UP000286642 UP000008912 UP000001519 UP000265180 UP000002281 UP000007819 UP000002277 UP000189706 UP000078542 UP000286640 UP000075809 UP000233220
UP000005203 UP000053097 UP000005447 UP000242457 UP000264820 UP000248484 UP000265300 UP000053105 UP000002494 UP000245320 UP000261500 UP000288706 UP000053825 UP000005215 UP000076502 UP000028990 UP000005226 UP000252040 UP000007303 UP000261640 UP000009136 UP000291000 UP000286641 UP000092461 UP000028760 UP000261480 UP000242638 UP000261540 UP000248483 UP000245340 UP000245341 UP000261380 UP000189704 UP000079721 UP000002852 UP000193380 UP000006813 UP000005205 UP000008227 UP000008225 UP000248481 UP000005640 UP000291020 UP000265120 UP000001038 UP000291022 UP000286642 UP000008912 UP000001519 UP000265180 UP000002281 UP000007819 UP000002277 UP000189706 UP000078542 UP000286640 UP000075809 UP000233220
Pfam
PF05458 Siva
SUPFAM
SSF57903
SSF57903
ProteinModelPortal
A0A3S2NTU8
A0A2H1VTR3
A0A2W1B8P2
A0A2A4JWK7
A0A0N1IK27
A0A194PPD8
+ More
S4NMZ4 A0A310SII5 A0A423T5T0 A0A0C9R2L2 A0A3Q8CBS8 O54926 Q3TFC3 A0A088ARN6 A0A026WT68 A0A286Y1S5 A0A2A3ELS2 A0A3Q2YAG8 A0A1E1X0W2 A0A2Y9T932 A0A340X8V0 A0A0N0U4X4 A4FTX4 A0A2U4B8G8 A0A3B3VE50 A0A444SCK9 A0A2U3V608 A0A0L7RDK0 I3MN94 A0A154PA52 A0A2R5LG96 P59692 A0A091CUX7 A0A3B5KGM8 A0A250YB36 A0A341CPU3 Q4RLN8 E9I8P0 A0A3Q3LDP7 V5HE68 G3X7B3 A0A452FIV7 A0A3Q7QA12 A0A1B0GLE0 A0A087XWY1 A0A3B3WND7 A0A3P9QBY1 A0A3B3RIN6 A0A2Y9NJS2 A0A2U3VL24 A0A2U3Y3U7 A0A3B5LRK6 A0A1B6C316 A0A1U7TIP2 A0A1S2ZF59 M3ZWX5 K9IWK9 A0A060WLI1 G5AWI6 A0A1B0CA90 A0A158NHF5 A0A315UYW8 M3VK34 A0A2R8MJ08 A0A2Y9G8P6 G3V439 A0A452IW01 A0A3P8UHT9 B7ZAZ2 B4DTY2 D2I109 H2MR92 A0A452QY36 A0A3Q7WD01 G1LMR9 G3V3U1 A0A2I2YVG9 F7HYP0 A0A3P9LR78 F7E257 J9JMK6 G3RB25 H2Q902 O15304 A0A1U7QL07 U3AQ02 A0A195D4F3 A0A3Q7U526 C4WUU7 A0A1A8HLF8 A0A151XFC9 A0A3Q7TTI3 A0A2K6S859 A0A3Q7TFG3 A0A2J8QLA6 A0A2J8QL97 A0A2J8SZ82 A0A2J8SZ79 A0A2J8QLI8 A0A1A8JQS9
S4NMZ4 A0A310SII5 A0A423T5T0 A0A0C9R2L2 A0A3Q8CBS8 O54926 Q3TFC3 A0A088ARN6 A0A026WT68 A0A286Y1S5 A0A2A3ELS2 A0A3Q2YAG8 A0A1E1X0W2 A0A2Y9T932 A0A340X8V0 A0A0N0U4X4 A4FTX4 A0A2U4B8G8 A0A3B3VE50 A0A444SCK9 A0A2U3V608 A0A0L7RDK0 I3MN94 A0A154PA52 A0A2R5LG96 P59692 A0A091CUX7 A0A3B5KGM8 A0A250YB36 A0A341CPU3 Q4RLN8 E9I8P0 A0A3Q3LDP7 V5HE68 G3X7B3 A0A452FIV7 A0A3Q7QA12 A0A1B0GLE0 A0A087XWY1 A0A3B3WND7 A0A3P9QBY1 A0A3B3RIN6 A0A2Y9NJS2 A0A2U3VL24 A0A2U3Y3U7 A0A3B5LRK6 A0A1B6C316 A0A1U7TIP2 A0A1S2ZF59 M3ZWX5 K9IWK9 A0A060WLI1 G5AWI6 A0A1B0CA90 A0A158NHF5 A0A315UYW8 M3VK34 A0A2R8MJ08 A0A2Y9G8P6 G3V439 A0A452IW01 A0A3P8UHT9 B7ZAZ2 B4DTY2 D2I109 H2MR92 A0A452QY36 A0A3Q7WD01 G1LMR9 G3V3U1 A0A2I2YVG9 F7HYP0 A0A3P9LR78 F7E257 J9JMK6 G3RB25 H2Q902 O15304 A0A1U7QL07 U3AQ02 A0A195D4F3 A0A3Q7U526 C4WUU7 A0A1A8HLF8 A0A151XFC9 A0A3Q7TTI3 A0A2K6S859 A0A3Q7TFG3 A0A2J8QLA6 A0A2J8QL97 A0A2J8SZ82 A0A2J8SZ79 A0A2J8QLI8 A0A1A8JQS9
Ontologies
KEGG
GO
PANTHER
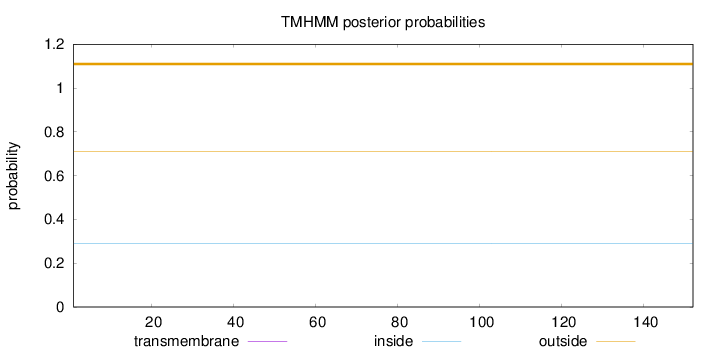
Topology
Subcellular location
Cytoplasm
In the nucleus, accumulates in dot-like structures. With evidence from 1 publications.
Nucleus In the nucleus, accumulates in dot-like structures. With evidence from 1 publications.
Nucleus In the nucleus, accumulates in dot-like structures. With evidence from 1 publications.
Length:
152
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00021
Exp number, first 60 AAs:
0
Total prob of N-in:
0.28865
outside
1 - 152
Population Genetic Test Statistics
Pi
169.111145
Theta
169.151408
Tajima's D
0.200988
CLR
13.724163
CSRT
0.424878756062197
Interpretation
Uncertain
