Gene
KWMTBOMO08581
Annotation
hypothetical_protein_OBRU01_10471?_partial_[Operophtera_brumata]
Location in the cell
Mitochondrial Reliability : 2.815
Sequence
CDS
ATGTTTCTGCGAAATATTTCACGTGCTATATACAATTGTAGTGGAAGGGCATCCTATGTAACAAAAGTACCAAAACCGGAGGTTCCAAAGAATCCTTTAGAGAAAGTCTATTCGGCATCTCAACAGAAAGATAATGGTATCATTTATGACAAAAAACCATTTAAAATAACTTTGGAAGCTGGTAAAACATATCACTGGTGTCTTTGCGGACGTTCCAAGAGACAGCCCTTATGCGACGGAACACACAAAGACATATTCCTTAAGGTCACGCAGAGGCCCATTAGATTCACAGTAGAAAAAACAAAGGATTATTGGCTGTGTAATTGTAAACAGACCAAGCTTAGACCTTTCTGTGATGGCACACACAAAAACAAAGATATACAAGAAGCTAATGTTAAAATTTAA
Protein
MFLRNISRAIYNCSGRASYVTKVPKPEVPKNPLEKVYSASQQKDNGIIYDKKPFKITLEAGKTYHWCLCGRSKRQPLCDGTHKDIFLKVTQRPIRFTVEKTKDYWLCNCKQTKLRPFCDGTHKNKDIQEANVKI
Summary
Uniprot
A0A0L7LDP0
A0A3S2M0M9
A0A2A4J499
A0A194PRC7
A0A0N1IIU0
I4DQF9
+ More
A0A1E1WKX7 A0A212F5F8 E2C0S0 E9IQ21 A0A151X5V0 A0A151IA16 A0A1W4XIY0 F4WB23 V5GCR5 A0A195EYK6 A0A158NRD6 A0A0J7L5C2 A0A151J617 A0A0K8TKA3 E2ADC8 A0A1Y1KD41 A0A0M9A219 A0A0A9YUN7 A0A0L7RCX7 A0A232EMR3 K7JHC2 W8BHZ0 A0A3B0JI12 A0A0C9R8H3 A0A182LHZ6 A0A182UMF6 A0A026WZN7 A0A224XQZ1 Q7Q987 A0A182PSZ5 A0A1I8PB00 A0A0V0G628 B4LK81 A0A1L8E810 A0A310SDY9 A0A1I8N3U5 A0A154P1Q3 A0A0M4EWJ2 A0A182WCU0 A0A2R7WZ12 A0A182M091 A0A069DMY2 A0A034WG57 A0A1A9WNV2 A0A0A1XMT8 A0A182JPA0 A0A1L8E128 A0A1L8E8D9 A0A1B0BFK5 A0A2M4C2J9 A0A2A3EI19 A0A2M4AN77 A0A1Q3FCC6 A0A182YBS2 A0A2J7QDM9 A0A182QKC6 A0A0K8UVA2 A0A1W4VFD4 B4P111 B4KNH5 B4MY45 B3MIN5 A0A088AM75 B3NB23 A0A1A9XAS7 B4J6K5 A0A067R5K8 Q0IFU5 Q292Y4 B4GC90 A0A0R3NKS2 W5J2J4 A0A0P4VMC6 A0A1A9V4T5 A0A1B0G2X3 A0A1A9ZX17 B4HQB7 A0A0L0BMJ8 A0A2M3ZBW3 B4QDC6 Q8SYQ8 A0A023EE04 A0A023EG46 A0A182GP96 J3JXK9 B0WC57 A0A1L8E8C0 A0A023EEN4 C4WV57 A0A182FE02 A0A1L8E7Q7 A0A182IU34 A0A182UB40
A0A1E1WKX7 A0A212F5F8 E2C0S0 E9IQ21 A0A151X5V0 A0A151IA16 A0A1W4XIY0 F4WB23 V5GCR5 A0A195EYK6 A0A158NRD6 A0A0J7L5C2 A0A151J617 A0A0K8TKA3 E2ADC8 A0A1Y1KD41 A0A0M9A219 A0A0A9YUN7 A0A0L7RCX7 A0A232EMR3 K7JHC2 W8BHZ0 A0A3B0JI12 A0A0C9R8H3 A0A182LHZ6 A0A182UMF6 A0A026WZN7 A0A224XQZ1 Q7Q987 A0A182PSZ5 A0A1I8PB00 A0A0V0G628 B4LK81 A0A1L8E810 A0A310SDY9 A0A1I8N3U5 A0A154P1Q3 A0A0M4EWJ2 A0A182WCU0 A0A2R7WZ12 A0A182M091 A0A069DMY2 A0A034WG57 A0A1A9WNV2 A0A0A1XMT8 A0A182JPA0 A0A1L8E128 A0A1L8E8D9 A0A1B0BFK5 A0A2M4C2J9 A0A2A3EI19 A0A2M4AN77 A0A1Q3FCC6 A0A182YBS2 A0A2J7QDM9 A0A182QKC6 A0A0K8UVA2 A0A1W4VFD4 B4P111 B4KNH5 B4MY45 B3MIN5 A0A088AM75 B3NB23 A0A1A9XAS7 B4J6K5 A0A067R5K8 Q0IFU5 Q292Y4 B4GC90 A0A0R3NKS2 W5J2J4 A0A0P4VMC6 A0A1A9V4T5 A0A1B0G2X3 A0A1A9ZX17 B4HQB7 A0A0L0BMJ8 A0A2M3ZBW3 B4QDC6 Q8SYQ8 A0A023EE04 A0A023EG46 A0A182GP96 J3JXK9 B0WC57 A0A1L8E8C0 A0A023EEN4 C4WV57 A0A182FE02 A0A1L8E7Q7 A0A182IU34 A0A182UB40
Pubmed
26227816
26354079
22651552
22118469
20798317
21282665
+ More
21719571 21347285 26369729 28004739 25401762 28648823 20075255 24495485 20966253 24508170 30249741 12364791 14747013 17210077 17994087 25315136 26334808 25348373 25830018 25244985 17550304 24845553 17510324 15632085 20920257 23761445 27129103 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 26483478 22516182
21719571 21347285 26369729 28004739 25401762 28648823 20075255 24495485 20966253 24508170 30249741 12364791 14747013 17210077 17994087 25315136 26334808 25348373 25830018 25244985 17550304 24845553 17510324 15632085 20920257 23761445 27129103 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24945155 26483478 22516182
EMBL
JTDY01001565
KOB73514.1
RSAL01000083
RVE48431.1
NWSH01003323
PCG66556.1
+ More
KQ459596 KPI95304.1 KQ459717 KPJ20320.1 AK404094 BAM20149.1 GDQN01008602 GDQN01003543 JAT82452.1 JAT87511.1 AGBW02010203 OWR48972.1 GL451846 EFN78464.1 GL764659 EFZ17376.1 KQ982494 KYQ55747.1 KQ978249 KYM96007.1 GL888056 EGI68578.1 GALX01006647 JAB61819.1 KQ981923 KYN32969.1 ADTU01023935 LBMM01000689 KMQ97748.1 KQ979928 KYN18610.1 GDAI01002949 JAI14654.1 GL438749 EFN68544.1 GEZM01087885 JAV58418.1 KQ435791 KOX74343.1 GBHO01022493 GBHO01006892 GBRD01003991 GBRD01003990 JAG21111.1 JAG36712.1 JAG61830.1 KQ414615 KOC68664.1 NNAY01003301 OXU19655.1 GAMC01010018 GAMC01010017 GAMC01010016 JAB96538.1 OUUW01000001 SPP73039.1 GBYB01003101 JAG72868.1 KK107054 QOIP01000004 EZA61497.1 RLU24132.1 GFTR01002912 JAW13514.1 AAAB01008905 EAA09698.3 GECL01002586 JAP03538.1 CH940648 EDW61672.1 GFDG01003931 JAV14868.1 KQ762882 OAD55387.1 KQ434796 KZC05791.1 CP012524 ALC42182.1 KK856074 PTY24788.1 AXCM01001770 GBGD01003476 JAC85413.1 GAKP01005650 JAC53302.1 GBXI01002419 JAD11873.1 GFDF01001654 JAV12430.1 GFDG01003930 JAV14869.1 JXJN01013534 GGFJ01010382 MBW59523.1 KZ288248 PBC31124.1 GGFK01008918 MBW42239.1 GFDL01009883 JAV25162.1 NEVH01015817 PNF26683.1 AXCN02001915 GDHF01021798 JAI30516.1 CM000157 EDW89085.1 CH933808 EDW08934.2 CH963894 EDW77034.1 CH902619 EDV38111.1 CH954177 EDV59788.1 CH916367 EDW02006.1 KK852946 KDR13421.1 CH477294 EAT44437.1 CM000071 EAL24727.2 CH479181 EDW31408.1 KRT01502.1 ADMH02002184 ETN57961.1 GDKW01002991 JAI53604.1 CCAG010015290 CH480816 EDW46656.1 JRES01001644 KNC21237.1 GGFM01005286 MBW26037.1 CM000362 CM002911 EDX05913.1 KMY91807.1 AY071376 AE013599 AAL48998.1 AAM70829.2 GAPW01005760 JAC07838.1 GAPW01005762 JAC07836.1 JXUM01170878 KQ578929 KXJ67776.1 BT127978 AEE62940.1 DS231885 EDS43209.1 GFDG01003885 JAV14914.1 GAPW01005761 JAC07837.1 ABLF02040229 AK341363 BAH71777.1 GFDG01004143 JAV14656.1
KQ459596 KPI95304.1 KQ459717 KPJ20320.1 AK404094 BAM20149.1 GDQN01008602 GDQN01003543 JAT82452.1 JAT87511.1 AGBW02010203 OWR48972.1 GL451846 EFN78464.1 GL764659 EFZ17376.1 KQ982494 KYQ55747.1 KQ978249 KYM96007.1 GL888056 EGI68578.1 GALX01006647 JAB61819.1 KQ981923 KYN32969.1 ADTU01023935 LBMM01000689 KMQ97748.1 KQ979928 KYN18610.1 GDAI01002949 JAI14654.1 GL438749 EFN68544.1 GEZM01087885 JAV58418.1 KQ435791 KOX74343.1 GBHO01022493 GBHO01006892 GBRD01003991 GBRD01003990 JAG21111.1 JAG36712.1 JAG61830.1 KQ414615 KOC68664.1 NNAY01003301 OXU19655.1 GAMC01010018 GAMC01010017 GAMC01010016 JAB96538.1 OUUW01000001 SPP73039.1 GBYB01003101 JAG72868.1 KK107054 QOIP01000004 EZA61497.1 RLU24132.1 GFTR01002912 JAW13514.1 AAAB01008905 EAA09698.3 GECL01002586 JAP03538.1 CH940648 EDW61672.1 GFDG01003931 JAV14868.1 KQ762882 OAD55387.1 KQ434796 KZC05791.1 CP012524 ALC42182.1 KK856074 PTY24788.1 AXCM01001770 GBGD01003476 JAC85413.1 GAKP01005650 JAC53302.1 GBXI01002419 JAD11873.1 GFDF01001654 JAV12430.1 GFDG01003930 JAV14869.1 JXJN01013534 GGFJ01010382 MBW59523.1 KZ288248 PBC31124.1 GGFK01008918 MBW42239.1 GFDL01009883 JAV25162.1 NEVH01015817 PNF26683.1 AXCN02001915 GDHF01021798 JAI30516.1 CM000157 EDW89085.1 CH933808 EDW08934.2 CH963894 EDW77034.1 CH902619 EDV38111.1 CH954177 EDV59788.1 CH916367 EDW02006.1 KK852946 KDR13421.1 CH477294 EAT44437.1 CM000071 EAL24727.2 CH479181 EDW31408.1 KRT01502.1 ADMH02002184 ETN57961.1 GDKW01002991 JAI53604.1 CCAG010015290 CH480816 EDW46656.1 JRES01001644 KNC21237.1 GGFM01005286 MBW26037.1 CM000362 CM002911 EDX05913.1 KMY91807.1 AY071376 AE013599 AAL48998.1 AAM70829.2 GAPW01005760 JAC07838.1 GAPW01005762 JAC07836.1 JXUM01170878 KQ578929 KXJ67776.1 BT127978 AEE62940.1 DS231885 EDS43209.1 GFDG01003885 JAV14914.1 GAPW01005761 JAC07837.1 ABLF02040229 AK341363 BAH71777.1 GFDG01004143 JAV14656.1
Proteomes
UP000037510
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000008237 UP000075809 UP000078542 UP000192223 UP000007755 UP000078541 UP000005205 UP000036403 UP000078492 UP000000311 UP000053105 UP000053825 UP000215335 UP000002358 UP000268350 UP000075882 UP000075903 UP000053097 UP000279307 UP000007062 UP000075885 UP000095300 UP000008792 UP000095301 UP000076502 UP000092553 UP000075920 UP000075883 UP000091820 UP000075881 UP000092460 UP000242457 UP000076408 UP000235965 UP000075886 UP000192221 UP000002282 UP000009192 UP000007798 UP000007801 UP000005203 UP000008711 UP000092443 UP000001070 UP000027135 UP000008820 UP000001819 UP000008744 UP000000673 UP000078200 UP000092444 UP000092445 UP000001292 UP000037069 UP000000304 UP000000803 UP000069940 UP000249989 UP000002320 UP000007819 UP000069272 UP000075880 UP000075902
UP000008237 UP000075809 UP000078542 UP000192223 UP000007755 UP000078541 UP000005205 UP000036403 UP000078492 UP000000311 UP000053105 UP000053825 UP000215335 UP000002358 UP000268350 UP000075882 UP000075903 UP000053097 UP000279307 UP000007062 UP000075885 UP000095300 UP000008792 UP000095301 UP000076502 UP000092553 UP000075920 UP000075883 UP000091820 UP000075881 UP000092460 UP000242457 UP000076408 UP000235965 UP000075886 UP000192221 UP000002282 UP000009192 UP000007798 UP000007801 UP000005203 UP000008711 UP000092443 UP000001070 UP000027135 UP000008820 UP000001819 UP000008744 UP000000673 UP000078200 UP000092444 UP000092445 UP000001292 UP000037069 UP000000304 UP000000803 UP000069940 UP000249989 UP000002320 UP000007819 UP000069272 UP000075880 UP000075902
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A0L7LDP0
A0A3S2M0M9
A0A2A4J499
A0A194PRC7
A0A0N1IIU0
I4DQF9
+ More
A0A1E1WKX7 A0A212F5F8 E2C0S0 E9IQ21 A0A151X5V0 A0A151IA16 A0A1W4XIY0 F4WB23 V5GCR5 A0A195EYK6 A0A158NRD6 A0A0J7L5C2 A0A151J617 A0A0K8TKA3 E2ADC8 A0A1Y1KD41 A0A0M9A219 A0A0A9YUN7 A0A0L7RCX7 A0A232EMR3 K7JHC2 W8BHZ0 A0A3B0JI12 A0A0C9R8H3 A0A182LHZ6 A0A182UMF6 A0A026WZN7 A0A224XQZ1 Q7Q987 A0A182PSZ5 A0A1I8PB00 A0A0V0G628 B4LK81 A0A1L8E810 A0A310SDY9 A0A1I8N3U5 A0A154P1Q3 A0A0M4EWJ2 A0A182WCU0 A0A2R7WZ12 A0A182M091 A0A069DMY2 A0A034WG57 A0A1A9WNV2 A0A0A1XMT8 A0A182JPA0 A0A1L8E128 A0A1L8E8D9 A0A1B0BFK5 A0A2M4C2J9 A0A2A3EI19 A0A2M4AN77 A0A1Q3FCC6 A0A182YBS2 A0A2J7QDM9 A0A182QKC6 A0A0K8UVA2 A0A1W4VFD4 B4P111 B4KNH5 B4MY45 B3MIN5 A0A088AM75 B3NB23 A0A1A9XAS7 B4J6K5 A0A067R5K8 Q0IFU5 Q292Y4 B4GC90 A0A0R3NKS2 W5J2J4 A0A0P4VMC6 A0A1A9V4T5 A0A1B0G2X3 A0A1A9ZX17 B4HQB7 A0A0L0BMJ8 A0A2M3ZBW3 B4QDC6 Q8SYQ8 A0A023EE04 A0A023EG46 A0A182GP96 J3JXK9 B0WC57 A0A1L8E8C0 A0A023EEN4 C4WV57 A0A182FE02 A0A1L8E7Q7 A0A182IU34 A0A182UB40
A0A1E1WKX7 A0A212F5F8 E2C0S0 E9IQ21 A0A151X5V0 A0A151IA16 A0A1W4XIY0 F4WB23 V5GCR5 A0A195EYK6 A0A158NRD6 A0A0J7L5C2 A0A151J617 A0A0K8TKA3 E2ADC8 A0A1Y1KD41 A0A0M9A219 A0A0A9YUN7 A0A0L7RCX7 A0A232EMR3 K7JHC2 W8BHZ0 A0A3B0JI12 A0A0C9R8H3 A0A182LHZ6 A0A182UMF6 A0A026WZN7 A0A224XQZ1 Q7Q987 A0A182PSZ5 A0A1I8PB00 A0A0V0G628 B4LK81 A0A1L8E810 A0A310SDY9 A0A1I8N3U5 A0A154P1Q3 A0A0M4EWJ2 A0A182WCU0 A0A2R7WZ12 A0A182M091 A0A069DMY2 A0A034WG57 A0A1A9WNV2 A0A0A1XMT8 A0A182JPA0 A0A1L8E128 A0A1L8E8D9 A0A1B0BFK5 A0A2M4C2J9 A0A2A3EI19 A0A2M4AN77 A0A1Q3FCC6 A0A182YBS2 A0A2J7QDM9 A0A182QKC6 A0A0K8UVA2 A0A1W4VFD4 B4P111 B4KNH5 B4MY45 B3MIN5 A0A088AM75 B3NB23 A0A1A9XAS7 B4J6K5 A0A067R5K8 Q0IFU5 Q292Y4 B4GC90 A0A0R3NKS2 W5J2J4 A0A0P4VMC6 A0A1A9V4T5 A0A1B0G2X3 A0A1A9ZX17 B4HQB7 A0A0L0BMJ8 A0A2M3ZBW3 B4QDC6 Q8SYQ8 A0A023EE04 A0A023EG46 A0A182GP96 J3JXK9 B0WC57 A0A1L8E8C0 A0A023EEN4 C4WV57 A0A182FE02 A0A1L8E7Q7 A0A182IU34 A0A182UB40
PDB
3TBN
E-value=2.7701e-18,
Score=219
Ontologies
KEGG
GO
PANTHER
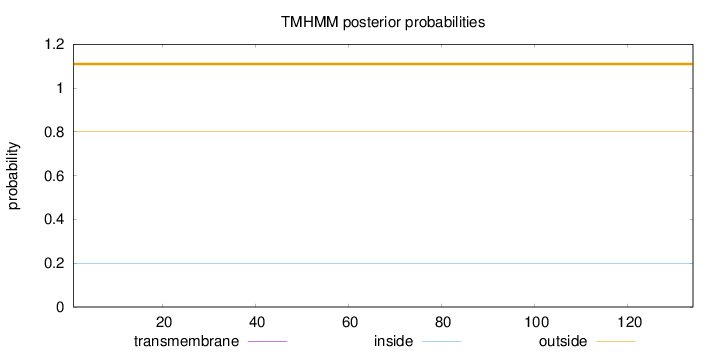
Topology
Length:
134
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.19839
outside
1 - 134
Population Genetic Test Statistics
Pi
150.069665
Theta
156.155485
Tajima's D
-0.401061
CLR
0.069873
CSRT
0.265886705664717
Interpretation
Uncertain
