Gene
KWMTBOMO08580
Pre Gene Modal
BGIBMGA007979
Annotation
PREDICTED:_cell_division_cycle_7-related_protein_kinase-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.533
Sequence
CDS
ATGTCCAGATCTAGGGGATTTGAAATAAGAGAAGGGTTACGTTCTCATAAAAAGCTCAAAGAAAATAAAAATGAAAATGAGACTTCTGATTCTTCTGAACGTGAACAAATATGTGAACTTTTATATAAGCTACCAAAACTGGACAAGATTTTTGAAGTGCACCGAAAGATAGGAGAAGGAACATTTAGTTCAGTGTATTTAGGCTCACTGAGACAGCATTCATCTTTGCTAGACAAAGACAAGCGTTGGTTTGCCATAAAACACTTGGTACCTACTGCACATCCAGCTCGGATTGAACACGAGTTAAGGTGTCTACAGGACATAGGAGGCAAACATCATGTGATTGGAGTGGAGCTCTGTCTTCGTAATCTCGACACTATTGTATTTGTAATGCCCTATCTACCTCATAAAAAGTTTTCTGAGTACGTGGGCGACATGGACGCGGCCGAGCTGCGCGAGTACATGCGCGCGCTGCTCGAGGCGCTCCGGCACGTGCACTCGTTCGGTGTCATACATCGAGACGTCAAACCCAGTAACTTTTTGTACGATCGAGAAAATAGGAGATATCAATTAGTAGACTTCGGCCTGGCTCAGCGACTGGTCCCCGGCACCGACGAGGAGCCCCCGGCGCCCCCCCGGTCCTCGGCCCCCCGCAAGCGTCCCCGGGAGGCCGAGGGCGCCACACCGGAAGCGAAGCGACTCGCACTTGACCTCTCACTGAAGACTAACGCTAGCAAAATATCTATTGTTGAAACTAATCCCATGCCGCTGTCTCCTAGAAGCAATGTCAAGAGAGCGGCGCCGTGCGCGTGCGCGGGTCGCGGCGCGGTGTGCGGCGCGTGCAGCGTGCGCCCCCCCGCCCGCGCCCCGCGCGCCGGCACGCAGGGCTTCCGCCCCCCCGAGGTGCTGCTGCGGTACGGCGCGCAGACCACCGCCGTGGACACGTGGGCGTGCGGCGTCGTGCTGTGCTCCGTGCTCACCGCGCGCTACCCGCTGCTGCGCGCCGCCGACGACCTCGCCGCCCTCGCCGAGCTCGCCGCGCTGCTCGGCACCCGCCCCCTGCAGCGCGCCGCCGCCGCCCTCGGTCGCCGTTTGATCACATCGTCCCCCCGACGCGGGCTGTGTCTCCGCAAGCTGTGCGCCCGCCTCCGCGGCTGTCCCCCCCCGGCCCCCGCGGCGCCGACCCCCGGCCCCCGGGCCCGCGCCCCCCGCCCCCTCCTCCCGGCGGCGTCGTGCTTGCACTGTCGGTGCCTCACGCACGAGTGTATTTGTCGAGACGAGGATAAAGACGCCGCCGCGGTGTCCCCGGGCCGCAGCGTGGCGGGGTTCCCGGACAGCGCGTTCTCGCTGTGCTGGCGGCTGCTGCAGCCGCTGCCGGCGCACAGGCTCTCCGCGGAGCAGGCCCTCCAACACGAGTTCTTCAACTCCTGA
Protein
MSRSRGFEIREGLRSHKKLKENKNENETSDSSEREQICELLYKLPKLDKIFEVHRKIGEGTFSSVYLGSLRQHSSLLDKDKRWFAIKHLVPTAHPARIEHELRCLQDIGGKHHVIGVELCLRNLDTIVFVMPYLPHKKFSEYVGDMDAAELREYMRALLEALRHVHSFGVIHRDVKPSNFLYDRENRRYQLVDFGLAQRLVPGTDEEPPAPPRSSAPRKRPREAEGATPEAKRLALDLSLKTNASKISIVETNPMPLSPRSNVKRAAPCACAGRGAVCGACSVRPPARAPRAGTQGFRPPEVLLRYGAQTTAVDTWACGVVLCSVLTARYPLLRAADDLAALAELAALLGTRPLQRAAAALGRRLITSSPRRGLCLRKLCARLRGCPPPAPAAPTPGPRARAPRPLLPAASCLHCRCLTHECICRDEDKDAAAVSPGRSVAGFPDSAFSLCWRLLQPLPAHRLSAEQALQHEFFNS
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
PDB
4F9C
E-value=4.69169e-45,
Score=457
Ontologies
GO
Topology
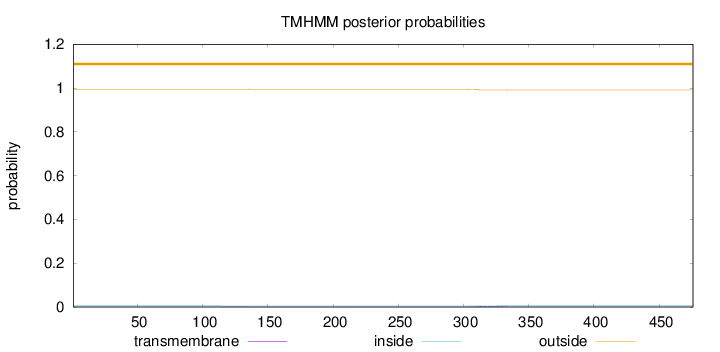
Length:
476
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1021
Exp number, first 60 AAs:
4e-05
Total prob of N-in:
0.00534
outside
1 - 476
Population Genetic Test Statistics
Pi
191.060689
Theta
156.89078
Tajima's D
0.712733
CLR
0.417148
CSRT
0.577421128943553
Interpretation
Uncertain
