Gene
KWMTBOMO08578 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007601
Annotation
PREDICTED:_delta-1-pyrroline-5-carboxylate_dehydrogenase?_mitochondrial_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 2.824
Sequence
CDS
ATGCTTACAATCTGGGCGAGAATCGGTCGCGTACGTTCGCTAGAGCGAACGTTGTCTAGCGTCGTAGACTTGCCGCAATTTCAGGATGGGAGCATCCATAACGAGCCTGTGTTGGGTTATCGTGCCGGAAGCCGGGAACGCCAAGACCTAATTGAGGAGCTGAAACGAACCGCTTCAGTAATTGAAGACGTACCAATTGTAATTGCCGGAAAAAATATTACAGAAGGCGAGCCACGATATCAGGTGATGCCCCATGATCACTCAAAAAAGATAGCTAAATATTACTATGCCAGTGAAAAAACAATTGAAAAGGCGATTCAAGCGTCTGTCGATGCTCAAACTCGTTGGGACCGCACTCCGCTGACTGAACGCGTAAAAATCTGGCAGACAGCAGCTGAGCTAATGGCCGGTGCGTATCGACAGAGCCTCAATGCTGCGACCATACTTGGACAGTCGAAATCAGTAATTCAAGCTGAAATTGATGCTGCAGCTGAACTTATTGATTTTTTCCGTTTTAATGTACACTTCCTGAAGGAAAATTCAAAATATCAACCTATTTCTGAGAGTCCCACAATAACCAAAAACTCTTTGAGATTTCGTGGCATAGATGGATTTATTGCAGCAATAAGTCCCTTCAATTTCACTGCCATTGGAGGCAACCTCGCATACACACCTGCACTTATGGGCAATGCAGTGTTATGGAAACCATCGGATACAGCTTTGTTGTCCAACTGGCGAATCTTCAATATTATGAGGGAAGCTGGTCTTCCAGATGGTGTTGTAAACTTTGTTCCATCCGATGGTCCTACATTTGGTCGAGTCATCACATCATCACCATACCTGGCTGGAATCAACTTCACAGGCTCTGTGCCCACTTTTAACTGGCTGTGGAATGAAGTAGGCAAGAATCTTAATACATATCGTAACTATCCCAGACTCATTGGAGAATGTGGAGGAAAGAATTACCACTTTGTACACCCCTCTGCTGATATACAAACTGTGGTTAGTGGCACCATCCGATCAGCATTTGAATTTTGTGGACAGAAGTGTTCAGCTTGCTCCAGAATATATATCCCTCAGTCTCTGTATGAACCAATTAAGACAGGATTACTTGCAGAAAGAGCTAAACTGAAGATTGGTGACCCTGCAGACTTTTCAGTGTTCACAGCGGCTGTCATTGATGACAAAGCTTTTGCTAGGATCACAGGATACATCCAAAGAGCCAAGAAGAATCCTAAGAACAAGATCTTGGGAGGTGGTGAGTTTGACAATAGCAAGGGCTACTTTGTGCAGCCTACCATCATTGAGACAACAGATCCGCATGACAAGCTCATGACAGAAGAGATCTTCGGTCCAGTGCTCACCATTTATGTATATAATGATAAAGATATTGATAAAACCATGGATTTAATTGGTACTTCCACAAAATTTGCTCTTACAGGAGCTATATTTGCTAAAGATCAAGTATTTTTGAAAAAAGCATATGAAAATCTCAAAATGACTGCTGGCAACTTCTATATCAATGACAAATCAACTGGATCTGTAGTCGGTCAGCAGCCATTTGGAGGTGGCCGTATGTCAGGTACTAATGACAAGGCAGGTGGTCCCAACTATGTCATGAGGTGGACAACTCCACAATCTATTAAGGAAACATTTGTTCCACTTACTGATATTGATTATCCATATATGAGAGAATGA
Protein
MLTIWARIGRVRSLERTLSSVVDLPQFQDGSIHNEPVLGYRAGSRERQDLIEELKRTASVIEDVPIVIAGKNITEGEPRYQVMPHDHSKKIAKYYYASEKTIEKAIQASVDAQTRWDRTPLTERVKIWQTAAELMAGAYRQSLNAATILGQSKSVIQAEIDAAAELIDFFRFNVHFLKENSKYQPISESPTITKNSLRFRGIDGFIAAISPFNFTAIGGNLAYTPALMGNAVLWKPSDTALLSNWRIFNIMREAGLPDGVVNFVPSDGPTFGRVITSSPYLAGINFTGSVPTFNWLWNEVGKNLNTYRNYPRLIGECGGKNYHFVHPSADIQTVVSGTIRSAFEFCGQKCSACSRIYIPQSLYEPIKTGLLAERAKLKIGDPADFSVFTAAVIDDKAFARITGYIQRAKKNPKNKILGGGEFDNSKGYFVQPTIIETTDPHDKLMTEEIFGPVLTIYVYNDKDIDKTMDLIGTSTKFALTGAIFAKDQVFLKKAYENLKMTAGNFYINDKSTGSVVGQQPFGGGRMSGTNDKAGGPNYVMRWTTPQSIKETFVPLTDIDYPYMRE
Summary
Similarity
Belongs to the aldehyde dehydrogenase family.
Uniprot
H9JDK4
A0A220K8K2
A0A2W1B9V2
A0A2A4J1B1
A0A0N1IH81
A0A194PPD3
+ More
A0A212F5G2 A0A1I8NER1 T1PE86 A0A182UUM8 A0A182XF32 A0A182LNJ8 Q7QA89 A0A182HVK0 A0A182TXD4 A0A182PG78 A0A0L0CEV1 A0A182W346 A0A182R8H8 A0A1I8QA61 A0A182YJQ4 A0A182KFH9 A0A023EU27 A0A0M4F1S2 A0A182H474 A0A2M3Z1V2 A0A182GZF5 A0A182SZD3 A0A182NCR2 A0A2M3Z1W0 A0A2M4BIN8 Q17A27 A0A182LZA6 A0A2M4BIQ5 A0A1J1IF54 A0A2M4A511 U5EY10 D2A4K8 A0A182QCA8 W5J9M2 T1EAL1 A0A182FP42 B0WKF4 A0A0T6BB18 A0A336MR06 B3MAI2 B4PI44 A0A1Q3FN06 A0A1Q3FMV8 A0A1Q3FMZ6 A0A0K8TMU3 A0A182JA00 B4NT61 B4N4H1 B3NJ08 A0A3B0J4I2 A0A1W4UE03 A0A084WQ65 Q9VNX4 B4L001 B4LC03 Q8T3P0 B4IAW9 A0A2P8YI55 N6TIF6 A0A2S2R7G8 A0A1A9XDC2 B4J0R3 Q2M042 B4GS53 A0A2S2PEX9 A0A1L8E0I3 A0A1L8E0C9 A0A067QNY3 A0A2U9K3B6 A0A1A9ZS83 A0A1B0BYU8 A0A1B0GD54 A0A2J7Q9V9 N6TBG3 V5I8H9 A0A1A9UQH1 A0A1B6GJ23 A0A1A9X107 G9FQ75 A0A1I9WLH5 A0A1Y1LI01 A0A0P4VJD2 A0A0C9Q928 A0A026WYS8 A0A1B6DE11 A0A069DVI3 A0A1W4XT86 A0A0J7L715 A0A224XMI1 K7IVK4 E2BU71 A0A232F6H4 A0A2P2HW63 A0A0V0G4C4
A0A212F5G2 A0A1I8NER1 T1PE86 A0A182UUM8 A0A182XF32 A0A182LNJ8 Q7QA89 A0A182HVK0 A0A182TXD4 A0A182PG78 A0A0L0CEV1 A0A182W346 A0A182R8H8 A0A1I8QA61 A0A182YJQ4 A0A182KFH9 A0A023EU27 A0A0M4F1S2 A0A182H474 A0A2M3Z1V2 A0A182GZF5 A0A182SZD3 A0A182NCR2 A0A2M3Z1W0 A0A2M4BIN8 Q17A27 A0A182LZA6 A0A2M4BIQ5 A0A1J1IF54 A0A2M4A511 U5EY10 D2A4K8 A0A182QCA8 W5J9M2 T1EAL1 A0A182FP42 B0WKF4 A0A0T6BB18 A0A336MR06 B3MAI2 B4PI44 A0A1Q3FN06 A0A1Q3FMV8 A0A1Q3FMZ6 A0A0K8TMU3 A0A182JA00 B4NT61 B4N4H1 B3NJ08 A0A3B0J4I2 A0A1W4UE03 A0A084WQ65 Q9VNX4 B4L001 B4LC03 Q8T3P0 B4IAW9 A0A2P8YI55 N6TIF6 A0A2S2R7G8 A0A1A9XDC2 B4J0R3 Q2M042 B4GS53 A0A2S2PEX9 A0A1L8E0I3 A0A1L8E0C9 A0A067QNY3 A0A2U9K3B6 A0A1A9ZS83 A0A1B0BYU8 A0A1B0GD54 A0A2J7Q9V9 N6TBG3 V5I8H9 A0A1A9UQH1 A0A1B6GJ23 A0A1A9X107 G9FQ75 A0A1I9WLH5 A0A1Y1LI01 A0A0P4VJD2 A0A0C9Q928 A0A026WYS8 A0A1B6DE11 A0A069DVI3 A0A1W4XT86 A0A0J7L715 A0A224XMI1 K7IVK4 E2BU71 A0A232F6H4 A0A2P2HW63 A0A0V0G4C4
Pubmed
19121390
28630352
28756777
26354079
22118469
25315136
+ More
20966253 12364791 14747013 17210077 26108605 25244985 24945155 26483478 17510324 18362917 19820115 20920257 23761445 17994087 17550304 26369729 22936249 18057021 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 23537049 15632085 23185243 24845553 27538518 28004739 27129103 24508170 30249741 26334808 20075255 20798317 28648823
20966253 12364791 14747013 17210077 26108605 25244985 24945155 26483478 17510324 18362917 19820115 20920257 23761445 17994087 17550304 26369729 22936249 18057021 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 23537049 15632085 23185243 24845553 27538518 28004739 27129103 24508170 30249741 26334808 20075255 20798317 28648823
EMBL
BABH01003355
MF319700
ASJ26414.1
KZ150375
PZC71115.1
NWSH01003734
+ More
PCG65865.1 KQ459717 KPJ20323.1 KQ459596 KPI95301.1 AGBW02010203 OWR48975.1 KA646994 KA649532 AFP61623.1 AAAB01008898 EAA09247.4 APCN01005788 JRES01000484 KNC30918.1 GAPW01000730 JAC12868.1 CP012525 ALC44945.1 JXUM01108965 JXUM01108966 JXUM01108967 KQ565279 KXJ71135.1 GGFM01001761 MBW22512.1 JXUM01021733 JXUM01021734 JXUM01021735 JXUM01021736 JXUM01021737 KQ560610 KXJ81633.1 GGFM01001742 MBW22493.1 GGFJ01003723 MBW52864.1 CH477340 EAT43137.1 AXCM01007092 GGFJ01003722 MBW52863.1 CVRI01000048 CRK98843.1 GGFK01002552 MBW35873.1 GANO01002252 JAB57619.1 KQ971344 EFA05239.1 AXCN02000334 ADMH02002085 ETN59444.1 GAMD01000943 JAB00648.1 DS231971 EDS29773.1 LJIG01002545 KRT84395.1 UFQT01002045 SSX32510.1 CH902618 EDV41269.1 CM000159 EDW95492.1 KRK02458.1 GFDL01006138 JAV28907.1 GFDL01006114 JAV28931.1 GFDL01006148 JAV28897.1 GDAI01001926 JAI15677.1 CH982756 CM002912 EDX15751.1 KMZ01109.1 CH964095 EDW79045.1 KRF98997.1 CH954178 EDV52725.1 KQS44401.1 OUUW01000002 SPP76415.1 ATLV01025273 KE525379 KFB52359.1 AE014296 BT050518 AAF51790.1 AAN12191.1 AAN12193.1 ACJ13225.1 ADV37595.1 AGB94889.1 CH933809 EDW19036.1 CH940647 EDW69803.1 KRF84586.1 KRF84587.1 KRF84588.1 KRF84589.1 AY094741 AAM11094.1 CH480826 EDW44432.1 PYGN01000578 PSN43931.1 APGK01036620 APGK01036621 KB740941 KB632409 ENN77583.1 ERL95167.1 GGMS01016437 MBY85640.1 CH916366 EDV97918.1 CH379069 EAL31094.1 KRT07862.1 KRT07863.1 CH479188 EDW40588.1 GGMR01015392 MBY28011.1 GFDF01001895 JAV12189.1 GFDF01001894 JAV12190.1 KK853123 KDR11043.1 MF446846 AWS20464.1 JXJN01022837 CCAG010002317 NEVH01016344 PNF25362.1 ENN77584.1 GALX01004655 JAB63811.1 GECZ01007348 JAS62421.1 JN187429 AET80385.2 KU932359 APA33995.1 GEZM01060188 JAV71206.1 GDKW01001508 JAI55087.1 GBYB01010783 JAG80550.1 KK107063 QOIP01000005 EZA61200.1 RLU22531.1 GEDC01013426 JAS23872.1 GBGD01000999 JAC87890.1 LBMM01000397 KMQ98458.1 GFTR01007205 JAW09221.1 GL450586 EFN80755.1 NNAY01000799 OXU26444.1 IACF01000224 LAB66023.1 GECL01003230 JAP02894.1
PCG65865.1 KQ459717 KPJ20323.1 KQ459596 KPI95301.1 AGBW02010203 OWR48975.1 KA646994 KA649532 AFP61623.1 AAAB01008898 EAA09247.4 APCN01005788 JRES01000484 KNC30918.1 GAPW01000730 JAC12868.1 CP012525 ALC44945.1 JXUM01108965 JXUM01108966 JXUM01108967 KQ565279 KXJ71135.1 GGFM01001761 MBW22512.1 JXUM01021733 JXUM01021734 JXUM01021735 JXUM01021736 JXUM01021737 KQ560610 KXJ81633.1 GGFM01001742 MBW22493.1 GGFJ01003723 MBW52864.1 CH477340 EAT43137.1 AXCM01007092 GGFJ01003722 MBW52863.1 CVRI01000048 CRK98843.1 GGFK01002552 MBW35873.1 GANO01002252 JAB57619.1 KQ971344 EFA05239.1 AXCN02000334 ADMH02002085 ETN59444.1 GAMD01000943 JAB00648.1 DS231971 EDS29773.1 LJIG01002545 KRT84395.1 UFQT01002045 SSX32510.1 CH902618 EDV41269.1 CM000159 EDW95492.1 KRK02458.1 GFDL01006138 JAV28907.1 GFDL01006114 JAV28931.1 GFDL01006148 JAV28897.1 GDAI01001926 JAI15677.1 CH982756 CM002912 EDX15751.1 KMZ01109.1 CH964095 EDW79045.1 KRF98997.1 CH954178 EDV52725.1 KQS44401.1 OUUW01000002 SPP76415.1 ATLV01025273 KE525379 KFB52359.1 AE014296 BT050518 AAF51790.1 AAN12191.1 AAN12193.1 ACJ13225.1 ADV37595.1 AGB94889.1 CH933809 EDW19036.1 CH940647 EDW69803.1 KRF84586.1 KRF84587.1 KRF84588.1 KRF84589.1 AY094741 AAM11094.1 CH480826 EDW44432.1 PYGN01000578 PSN43931.1 APGK01036620 APGK01036621 KB740941 KB632409 ENN77583.1 ERL95167.1 GGMS01016437 MBY85640.1 CH916366 EDV97918.1 CH379069 EAL31094.1 KRT07862.1 KRT07863.1 CH479188 EDW40588.1 GGMR01015392 MBY28011.1 GFDF01001895 JAV12189.1 GFDF01001894 JAV12190.1 KK853123 KDR11043.1 MF446846 AWS20464.1 JXJN01022837 CCAG010002317 NEVH01016344 PNF25362.1 ENN77584.1 GALX01004655 JAB63811.1 GECZ01007348 JAS62421.1 JN187429 AET80385.2 KU932359 APA33995.1 GEZM01060188 JAV71206.1 GDKW01001508 JAI55087.1 GBYB01010783 JAG80550.1 KK107063 QOIP01000005 EZA61200.1 RLU22531.1 GEDC01013426 JAS23872.1 GBGD01000999 JAC87890.1 LBMM01000397 KMQ98458.1 GFTR01007205 JAW09221.1 GL450586 EFN80755.1 NNAY01000799 OXU26444.1 IACF01000224 LAB66023.1 GECL01003230 JAP02894.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000095301
+ More
UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000075885 UP000037069 UP000075920 UP000075900 UP000095300 UP000076408 UP000075881 UP000092553 UP000069940 UP000249989 UP000075901 UP000075884 UP000008820 UP000075883 UP000183832 UP000007266 UP000075886 UP000000673 UP000069272 UP000002320 UP000007801 UP000002282 UP000075880 UP000000304 UP000007798 UP000008711 UP000268350 UP000192221 UP000030765 UP000000803 UP000009192 UP000008792 UP000001292 UP000245037 UP000019118 UP000030742 UP000092443 UP000001070 UP000001819 UP000008744 UP000027135 UP000092445 UP000092460 UP000092444 UP000235965 UP000078200 UP000091820 UP000053097 UP000279307 UP000192223 UP000036403 UP000002358 UP000008237 UP000215335
UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000075885 UP000037069 UP000075920 UP000075900 UP000095300 UP000076408 UP000075881 UP000092553 UP000069940 UP000249989 UP000075901 UP000075884 UP000008820 UP000075883 UP000183832 UP000007266 UP000075886 UP000000673 UP000069272 UP000002320 UP000007801 UP000002282 UP000075880 UP000000304 UP000007798 UP000008711 UP000268350 UP000192221 UP000030765 UP000000803 UP000009192 UP000008792 UP000001292 UP000245037 UP000019118 UP000030742 UP000092443 UP000001070 UP000001819 UP000008744 UP000027135 UP000092445 UP000092460 UP000092444 UP000235965 UP000078200 UP000091820 UP000053097 UP000279307 UP000192223 UP000036403 UP000002358 UP000008237 UP000215335
Interpro
IPR016161
Ald_DH/histidinol_DH
+ More
IPR016163 Ald_DH_C
IPR016160 Ald_DH_CS_CYS
IPR005931 P5CDH/ALDH4A1
IPR016162 Ald_DH_N
IPR015590 Aldehyde_DH_dom
IPR029510 Ald_DH_CS_GLU
IPR036517 FF_domain_sf
IPR006638 Elp3/MiaB/NifB
IPR002713 FF_domain
IPR006463 MiaB_methiolase
IPR005839 Methylthiotransferase
IPR001202 WW_dom
IPR023404 rSAM_horseshoe
IPR013848 Methylthiotransferase_N
IPR020612 Methylthiotransferase_CS
IPR038135 Methylthiotransferase_N_sf
IPR036020 WW_dom_sf
IPR007197 rSAM
IPR016163 Ald_DH_C
IPR016160 Ald_DH_CS_CYS
IPR005931 P5CDH/ALDH4A1
IPR016162 Ald_DH_N
IPR015590 Aldehyde_DH_dom
IPR029510 Ald_DH_CS_GLU
IPR036517 FF_domain_sf
IPR006638 Elp3/MiaB/NifB
IPR002713 FF_domain
IPR006463 MiaB_methiolase
IPR005839 Methylthiotransferase
IPR001202 WW_dom
IPR023404 rSAM_horseshoe
IPR013848 Methylthiotransferase_N
IPR020612 Methylthiotransferase_CS
IPR038135 Methylthiotransferase_N_sf
IPR036020 WW_dom_sf
IPR007197 rSAM
ProteinModelPortal
H9JDK4
A0A220K8K2
A0A2W1B9V2
A0A2A4J1B1
A0A0N1IH81
A0A194PPD3
+ More
A0A212F5G2 A0A1I8NER1 T1PE86 A0A182UUM8 A0A182XF32 A0A182LNJ8 Q7QA89 A0A182HVK0 A0A182TXD4 A0A182PG78 A0A0L0CEV1 A0A182W346 A0A182R8H8 A0A1I8QA61 A0A182YJQ4 A0A182KFH9 A0A023EU27 A0A0M4F1S2 A0A182H474 A0A2M3Z1V2 A0A182GZF5 A0A182SZD3 A0A182NCR2 A0A2M3Z1W0 A0A2M4BIN8 Q17A27 A0A182LZA6 A0A2M4BIQ5 A0A1J1IF54 A0A2M4A511 U5EY10 D2A4K8 A0A182QCA8 W5J9M2 T1EAL1 A0A182FP42 B0WKF4 A0A0T6BB18 A0A336MR06 B3MAI2 B4PI44 A0A1Q3FN06 A0A1Q3FMV8 A0A1Q3FMZ6 A0A0K8TMU3 A0A182JA00 B4NT61 B4N4H1 B3NJ08 A0A3B0J4I2 A0A1W4UE03 A0A084WQ65 Q9VNX4 B4L001 B4LC03 Q8T3P0 B4IAW9 A0A2P8YI55 N6TIF6 A0A2S2R7G8 A0A1A9XDC2 B4J0R3 Q2M042 B4GS53 A0A2S2PEX9 A0A1L8E0I3 A0A1L8E0C9 A0A067QNY3 A0A2U9K3B6 A0A1A9ZS83 A0A1B0BYU8 A0A1B0GD54 A0A2J7Q9V9 N6TBG3 V5I8H9 A0A1A9UQH1 A0A1B6GJ23 A0A1A9X107 G9FQ75 A0A1I9WLH5 A0A1Y1LI01 A0A0P4VJD2 A0A0C9Q928 A0A026WYS8 A0A1B6DE11 A0A069DVI3 A0A1W4XT86 A0A0J7L715 A0A224XMI1 K7IVK4 E2BU71 A0A232F6H4 A0A2P2HW63 A0A0V0G4C4
A0A212F5G2 A0A1I8NER1 T1PE86 A0A182UUM8 A0A182XF32 A0A182LNJ8 Q7QA89 A0A182HVK0 A0A182TXD4 A0A182PG78 A0A0L0CEV1 A0A182W346 A0A182R8H8 A0A1I8QA61 A0A182YJQ4 A0A182KFH9 A0A023EU27 A0A0M4F1S2 A0A182H474 A0A2M3Z1V2 A0A182GZF5 A0A182SZD3 A0A182NCR2 A0A2M3Z1W0 A0A2M4BIN8 Q17A27 A0A182LZA6 A0A2M4BIQ5 A0A1J1IF54 A0A2M4A511 U5EY10 D2A4K8 A0A182QCA8 W5J9M2 T1EAL1 A0A182FP42 B0WKF4 A0A0T6BB18 A0A336MR06 B3MAI2 B4PI44 A0A1Q3FN06 A0A1Q3FMV8 A0A1Q3FMZ6 A0A0K8TMU3 A0A182JA00 B4NT61 B4N4H1 B3NJ08 A0A3B0J4I2 A0A1W4UE03 A0A084WQ65 Q9VNX4 B4L001 B4LC03 Q8T3P0 B4IAW9 A0A2P8YI55 N6TIF6 A0A2S2R7G8 A0A1A9XDC2 B4J0R3 Q2M042 B4GS53 A0A2S2PEX9 A0A1L8E0I3 A0A1L8E0C9 A0A067QNY3 A0A2U9K3B6 A0A1A9ZS83 A0A1B0BYU8 A0A1B0GD54 A0A2J7Q9V9 N6TBG3 V5I8H9 A0A1A9UQH1 A0A1B6GJ23 A0A1A9X107 G9FQ75 A0A1I9WLH5 A0A1Y1LI01 A0A0P4VJD2 A0A0C9Q928 A0A026WYS8 A0A1B6DE11 A0A069DVI3 A0A1W4XT86 A0A0J7L715 A0A224XMI1 K7IVK4 E2BU71 A0A232F6H4 A0A2P2HW63 A0A0V0G4C4
PDB
4LH3
E-value=6.09674e-180,
Score=1621
Ontologies
PATHWAY
GO
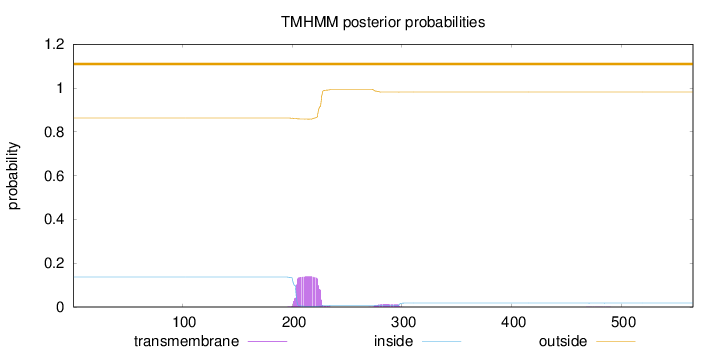
Topology
Length:
565
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.43137000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.13732
outside
1 - 565
Population Genetic Test Statistics
Pi
172.045747
Theta
198.413366
Tajima's D
-0.309515
CLR
0.058655
CSRT
0.291385430728464
Interpretation
Uncertain
